 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA38997 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 304aa MW: 34504.2 Da PI: 8.8813 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 95.4 | 2.5e-30 | 9 | 58 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krienk+nrqvtfskRrng+lKKA+ELSvLCdae+a+iifss+gkl e+
Tp57577_TGAC_v2_mRNA38997 9 KRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEIALIIFSSRGKLSEFG 58
79**********************************************95 PP
| |||||||
| 2 | K-box | 25.2 | 6.9e-10 | 78 | 123 | 4 | 48 |
K-box 4 ssgksleeakaeslqqelakLkkeienLqreqRh.llGedLesLsl 48
s++ + +e+++++++qe++kLk+++e+Lq++q + +G L+sL++
Tp57577_TGAC_v2_mRNA38997 78 SQNDNDNEHETQNWYQEISKLKAKYESLQKSQSNsNKGFRLQSLTI 123
4455578899*******************88877258999999987 PP
| |||||||
| 3 | K-box | 63.2 | 9.5e-22 | 144 | 196 | 36 | 88 |
K-box 36 RhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelq 88
R+llGedL++L++keLq+Le+qLe +l + R++K+++++eq+eel+kk ++ q
Tp57577_TGAC_v2_mRNA38997 144 RQLLGEDLGPLNIKELQNLEKQLEGALAQARQRKTQIMIEQMEELRKKVRNSQ 196
9************************************************9976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 8.2E-41 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 32.576 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 7.38E-42 | 2 | 80 | No hit | No description |
| SuperFamily | SSF55455 | 2.75E-33 | 2 | 92 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.3E-31 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.4E-25 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.3E-31 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.3E-31 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 13.331 | 88 | 232 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 2.9E-16 | 143 | 196 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009553 | Biological Process | embryo sac development | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010094 | Biological Process | specification of carpel identity | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048455 | Biological Process | stamen formation | ||||
| GO:0048459 | Biological Process | floral whorl structural organization | ||||
| GO:0048509 | Biological Process | regulation of meristem development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0080112 | Biological Process | seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 304 aa Download sequence Send to blast |
MGRGRVELKR IENKINRQVT FSKRRNGLLK KAYELSVLCD AEIALIIFSS RGKLSEFGSS 60 TSGIGKTLER YQRCSYTSQN DNDNEHETQN WYQEISKLKA KYESLQKSQS NSNKGFRLQS 120 LTIEITLGLS LCWVGPNCHV NYFRQLLGED LGPLNIKELQ NLEKQLEGAL AQARQRKTQI 180 MIEQMEELRK KVRNSQVPTN LGILPSSIIY LELHLGEINK QLRFKLESDG FNLKAYESMW 240 NTTNSASVAG GGNFPFQPSE TNPMDCQPEP FLQIGYQNYV QAEQSSAPKN MVGETSFIQG 300 WML* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 6e-19 | 1 | 71 | 1 | 69 | MEF2C |
| 5f28_B | 6e-19 | 1 | 71 | 1 | 69 | MEF2C |
| 5f28_C | 6e-19 | 1 | 71 | 1 | 69 | MEF2C |
| 5f28_D | 6e-19 | 1 | 71 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
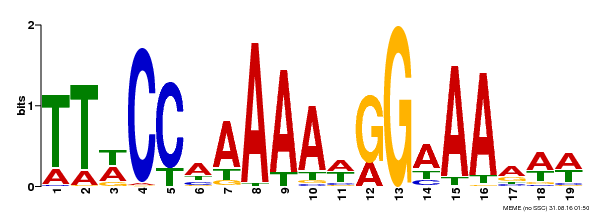
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA38997 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC147014 | 2e-62 | AC147014.29 Medicago truncatula clone mth2-164l15, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003623805.2 | 1e-158 | agamous-like MADS-box protein AGL6 | ||||
| Swissprot | Q8LLR1 | 1e-118 | MADS3_VITVI; Agamous-like MADS-box protein MADS3 | ||||
| TrEMBL | G7L1F4 | 1e-157 | G7L1F4_MEDTR; MADS-box transcription factor | ||||
| STRING | AES80023 | 1e-152 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF229 | 32 | 178 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 1e-99 | AGAMOUS-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA38997 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



