 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA39106 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 813aa MW: 89272 Da PI: 6.4298 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.6 | 7e-21 | 127 | 182 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Tp57577_TGAC_v2_mRNA39106 127 KKRYHRHTPQQIQELESMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 182
688999***********************************************999 PP
| |||||||
| 2 | START | 198.1 | 4e-62 | 325 | 551 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS........SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-T CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.......dsgealrasgvvdmvlallveellddkeqWde 74
la+ a++elvk+a+ +ep+W +s+ e++n +e+++++++s + +ea+r+sgvv+ ++ lve+l+d++ +W+e
Tp57577_TGAC_v2_mRNA39106 325 LALVAMDELVKMAQTNEPLWIRSVeggrEILNHEEYTRTISTSSCiglkpngFVSEASRESGVVIINSLALVETLMDSN-RWSE 407
67889***********************************99888**********************************.**** PP
T-S....EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--..-TTSEE-E CS
START 75 tla....kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.sssvvRae 146
+++ + +t evissg galqlm+aelq+lsplvp R + f+R+++q+ +g+w++vdvS+d ++ + s + ++
Tp57577_TGAC_v2_mRNA39106 408 MFPcviaRTSTNEVISSGingtrnGALQLMQAELQVLSPLVPvRLVNFLRFCKQHAEGVWAVVDVSIDTITQTSAvASTFLNCR 491
*********************************************************************9999999******** PP
ESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 147 llpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+lpSg+l+++++ng+skvtwveh++++++++h+l+r+l++ g+ +ga++wvatlqrqce+
Tp57577_TGAC_v2_mRNA39106 492 KLPSGFLVQDMPNGYSKVTWVEHTEYEESQVHQLYRPLLSLGMGFGAQRWVATLQRQCEC 551
**********************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.71E-20 | 117 | 184 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-22 | 119 | 184 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.297 | 124 | 184 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.5E-18 | 125 | 188 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.13E-18 | 126 | 184 | No hit | No description |
| Pfam | PF00046 | 2.1E-18 | 127 | 182 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 159 | 182 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 42.613 | 315 | 554 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.93E-32 | 317 | 551 | No hit | No description |
| CDD | cd08875 | 1.78E-118 | 319 | 550 | No hit | No description |
| SMART | SM00234 | 1.3E-46 | 324 | 551 | IPR002913 | START domain |
| Pfam | PF01852 | 6.2E-54 | 325 | 551 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.93E-23 | 579 | 805 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 813 aa Download sequence Send to blast |
MSFGDNKSGC GESAKISSVS DIAFSNGTNH NNNYRIIMPS SAISQPHLST PTLVKSMFNS 60 PGLSLSLQTN MEGQGGGGDM SRLMAESFHG LRRNREEEEE EHESRSGGSD NIDGVSGDDA 120 ADNPPRKKRY HRHTPQQIQE LESMFKECPH PDEKQRLELS KRLCLETRQV KFWFQNRRTQ 180 MKTQLERHEN SLLRQANDKL RVENMSIREA MRNPMCSNCG GPTIINEISL EEQHLRIENA 240 RLKDDLERVC TLASKFLGRP ITSLPNSSLE FSFVGDHSNT LPSTLPLGQD FGMVSISPPS 300 TIIATNSTSS NGFDRSIERS MLLELALVAM DELVKMAQTN EPLWIRSVEG GREILNHEEY 360 TRTISTSSCI GLKPNGFVSE ASRESGVVII NSLALVETLM DSNRWSEMFP CVIARTSTNE 420 VISSGINGTR NGALQLMQAE LQVLSPLVPV RLVNFLRFCK QHAEGVWAVV DVSIDTITQT 480 SAVASTFLNC RKLPSGFLVQ DMPNGYSKVT WVEHTEYEES QVHQLYRPLL SLGMGFGAQR 540 WVATLQRQCE CLAILMSSAL PSREQSAISS GGRRSMLKLA QRMTNNFCAG VCASTVHKWN 600 KLNAGNVGED VRVMTRKSVD DPGEPSGVVL SAATSVWLPA SQQKVFNFLR NERLRSEWDI 660 LSNGGPMQEM AHIAKGHDHG NCVSLLRASG NNSSQSSMLI LQETCTDASG SLVVYAPVDI 720 PAMHVVMNGG DSAYVALLPS GFAILPDGYS DDSDNQRGGL QHRANGSLLT VAFQILVNSL 780 PTAKLTVESV ETVNNLISCT VQKIKAALQC ES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
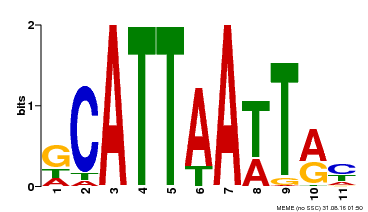
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA39106 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC148764 | 0.0 | AC148764.16 Medicago truncatula clone mth2-30o12, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003623826.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A2Z6N8C0 | 0.0 | A0A2Z6N8C0_TRISU; Uncharacterized protein | ||||
| STRING | AES80044 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA39106 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



