 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA41565 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 455aa MW: 49806.1 Da PI: 6.7122 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.2 | 1.1e-10 | 195 | 236 | 5 | 46 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLk 46
k rr+++NReAA++sR+RKka++++Le+ L++ ++L+
Tp57577_TGAC_v2_mRNA41565 195 KTLRRLAQNREAAKKSRLRKKAYVQQLETSRLRLSNLEQDLQ 236
6789************************98888887777665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.4E-7 | 187 | 237 | No hit | No description |
| SMART | SM00338 | 2.9E-6 | 191 | 253 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.013 | 193 | 237 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.69E-6 | 195 | 239 | No hit | No description |
| Pfam | PF00170 | 2.7E-8 | 195 | 236 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 198 | 213 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 8.3E-27 | 297 | 372 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 455 aa Download sequence Send to blast |
MASQRIGEIG LSESGPSSQH VPYGVLHGIT NSTSTLMNQG SAFDFGELEE AIVLQGIKIR 60 NDEGKSSALF TSKPAATLEM FPSWPIRFQQ SPRVVASLTS PSLTIPYGGS KSGGESTDSG 120 STELQFETDS PISVKASSSD HHNNHNHVFD QQLQQETATD DSLRTGTSQN QSKAKSHNEK 180 KKGAVSTSEK TLDPKTLRRL AQNREAAKKS RLRKKAYVQQ LETSRLRLSN LEQDLQRARS 240 QVCVRSSAVK MHAVSTYKSS DLDQTGLFLG CGGGNISPGA AMFDMEYARW LEEDQRQLAE 300 LRAGLQAALP DNELRVLVDG YLYHYDELFR LKGVAVKSDV FHLIKGIWAS PAERPFIWIG 360 GFKPTELITM LTQQLEPLAE QQLAGIMELG KSAQQAEEAL SKGHEQLHHA IVDTIAGGDV 420 IDGVQQMVAA MARISNLEGF VYQVNKLIIS TLYM* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
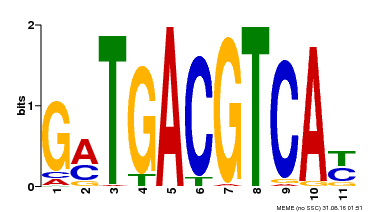
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA41565 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003606112.1 | 0.0 | transcription factor TGA9 | ||||
| Refseq | XP_024637666.1 | 0.0 | transcription factor TGA9 | ||||
| Swissprot | Q93XM6 | 1e-134 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | G7JIE0 | 0.0 | G7JIE0_MEDTR; BZIP family transcription factor | ||||
| STRING | AES88309 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4555 | 33 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.3 | 1e-126 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA41565 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



