 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA4230 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 289aa MW: 32458.9 Da PI: 6.2547 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 108.7 | 2.9e-34 | 141 | 198 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s+fprsYYrCts +C+vkk+vers +dp++v++tYeg+H+h+
Tp57577_TGAC_v2_mRNA4230 141 EDGYRWRKYGQKAVKNSPFPRSYYRCTSVSCNVKKHVERSLNDPTIVVTTYEGKHTHP 198
8********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF038130 | 6.2E-107 | 2 | 288 | IPR017396 | PWRKY transcription factor, group IIc |
| Gene3D | G3DSA:2.20.25.80 | 3.6E-35 | 125 | 200 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.09E-29 | 132 | 200 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.374 | 135 | 200 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.6E-40 | 140 | 199 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.3E-26 | 141 | 198 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009624 | Biological Process | response to nematode | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MEEDYEIGSY FPSYSLSSVF DLSEDKSSSV GGFMELLGVQ NMSSMFDYPV DDAVVKETSL 60 EKKECSEVLN SNSISQQPAT PNSSSISSAS SEAINDEQTK TVDQANNQLN KQLKAKKTTN 120 QKRQREARIA FMTKSEVDHL EDGYRWRKYG QKAVKNSPFP RSYYRCTSVS CNVKKHVERS 180 LNDPTIVVTT YEGKHTHPNP VMARSSSVHA GSLLPSPECT TNFVSNQNYN MSQYYNQQRQ 240 QALFSNLSSL GFTSKNDAQE RSLCNNPRLM DNGLLQDVVP SHMFKEEE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-28 | 133 | 200 | 10 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-28 | 133 | 200 | 10 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00069 | PBM | Transfer from AT2G47260 | Download |
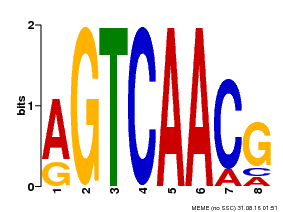
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA4230 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT146334 | 0.0 | BT146334.1 Medicago truncatula clone JCVI-FLMt-7D17 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003625994.2 | 1e-140 | probable WRKY transcription factor 23 | ||||
| Swissprot | O22900 | 5e-62 | WRK23_ARATH; WRKY transcription factor 23 | ||||
| TrEMBL | A0A2K3MUR6 | 0.0 | A0A2K3MUR6_TRIPR; WRKY transcription factor | ||||
| STRING | AES82212 | 1e-140 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1002 | 34 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47260.1 | 4e-53 | WRKY DNA-binding protein 23 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA4230 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



