 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA4562 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 308aa MW: 34434.3 Da PI: 6.6172 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.1 | 3.6e-17 | 14 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT.-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGgg.tWktIartmgkgRtlkqcksrwqkyl 48
+g+WTteEde+lv+ +k+ Gg+ +W++ ++ g++R++k+c++rw +yl
Tp57577_TGAC_v2_mRNA4562 14 KGPWTTEEDEILVNFIKKNGGHgSWRSLPKLAGLRRCGKSCRLRWTNYL 62
79*******************9*************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 47.1 | 5.6e-15 | 68 | 113 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T++E++l +++++ lG++ W++Ia+ + gRt++++k+ w+++l
Tp57577_TGAC_v2_mRNA4562 68 RGPFTPQEEKLVIQLHAILGNR-WAAIASQLA-GRTDNEIKNLWNTHL 113
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.336 | 9 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.74E-28 | 10 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.7E-12 | 13 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-14 | 14 | 62 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.59E-9 | 16 | 62 | No hit | No description |
| PROSITE profile | PS51294 | 24.454 | 63 | 117 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-25 | 65 | 118 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.0E-14 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.3E-14 | 68 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.88E-10 | 70 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 308 aa Download sequence Send to blast |
MGRTPCCDKM GLKKGPWTTE EDEILVNFIK KNGGHGSWRS LPKLAGLRRC GKSCRLRWTN 60 YLRPDIKRGP FTPQEEKLVI QLHAILGNRW AAIASQLAGR TDNEIKNLWN THLKKRLVCM 120 GIDPQTHEPI ASSHYYPLSK SHSSIATRHM AQWESARLEA EARLSKDSSL FNNNNIPSFA 180 SNKTDSDSDY FLKIWNSEVG QSFRSVRKSD DGDDDDDKTR CISSMSQEGS SCNNTKCGSV 240 SAITTITDLA SSTLASNVKE DFEWRNYKSF DIGCDDSSSS NDLEDSSDTA LQLLLDFPSN 300 NDMSFLE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-26 | 12 | 117 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may play a role in flower development by repressing ANT (PubMed:19232308). Regulates the transition of meristem identity from vegetative growth to flowering. Acts downstream of LFY and upstream of AP1. Directly activates AP1 to promote floral fate. Together with LFY and AP1 may constitute a regulatory network that contributes to an abrupt and robust meristem identity transition (PubMed:21750030). {ECO:0000269|PubMed:19232308, ECO:0000269|PubMed:21750030}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00114 | ampDAP | Transfer from AT3G61250 | Download |
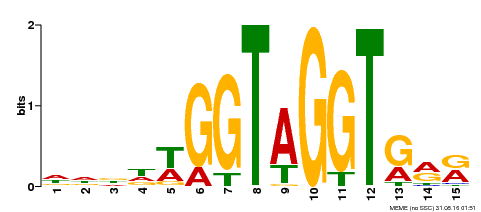
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA4562 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013444869.1 | 0.0 | transcription factor MYB41 | ||||
| Swissprot | Q9M2D9 | 1e-120 | MYB17_ARATH; Transcription factor MYB17 | ||||
| TrEMBL | A0A2K3NBN9 | 0.0 | A0A2K3NBN9_TRIPR; Protein ODORANT1-like | ||||
| STRING | AES75206 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2331 | 32 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61250.1 | 1e-114 | myb domain protein 17 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA4562 |



