 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA8623 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 306aa MW: 35370 Da PI: 6.8656 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 87.8 | 5.7e-28 | 19 | 69 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien rqvtfskRr+g+lKKA+EL +LCdaevaviifs+tgkl+e+ss
Tp57577_TGAC_v2_mRNA8623 19 KRIENANSRQVTFSKRRTGLLKKAQELAILCDAEVAVIIFSNTGKLFEFSS 69
79***********************************************96 PP
| |||||||
| 2 | K-box | 60.2 | 8.4e-21 | 150 | 230 | 20 | 100 |
K-box 20 elakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
++ Lk+ei +L+++q +llG+dL+ L lkeLq+Leqq++++l +++++K+ell+eq+e+++ +e+++ en++Lr+++ee
Tp57577_TGAC_v2_mRNA8623 150 MVEILKDEIAKLETNQLRLLGKDLTGLGLKELQHLEQQINEGLLSVKERKEELLMEQLEQSRIQEQKAMLENETLRRQVEE 230
5778**************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 32.492 | 11 | 71 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 7.9E-42 | 11 | 70 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 3.01E-29 | 11 | 80 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 2.32E-37 | 12 | 114 | No hit | No description |
| PRINTS | PR00404 | 3.6E-29 | 13 | 33 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 13 | 67 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 9.9E-26 | 20 | 67 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 3.6E-29 | 33 | 48 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 3.6E-29 | 48 | 69 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 12.989 | 144 | 234 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 5.4E-15 | 151 | 228 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 306 aa Download sequence Send to blast |
LGFLCFEKEE MGRGKIEIKR IENANSRQVT FSKRRTGLLK KAQELAILCD AEVAVIIFSN 60 TGKLFEFSSS GYVFEVFEFF DFIACLFYFV CEFCLLIMKR TLSRYNKCTD SSESASVEYK 120 TEILVIYSLI CCLILMSINK YFLQKDDTKM VEILKDEIAK LETNQLRLLG KDLTGLGLKE 180 LQHLEQQINE GLLSVKERKE ELLMEQLEQS RIQEQKAMLE NETLRRQVEE LRCLFPIAER 240 VMPTYLQYHH VERKNSFVDN SVKCPSFARN CANDKADSDT TLHLGLPTDV RNKKTPEKKK 300 ISNDS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 1e-17 | 11 | 70 | 1 | 60 | MEF2C |
| 5f28_B | 1e-17 | 11 | 70 | 1 | 60 | MEF2C |
| 5f28_C | 1e-17 | 11 | 70 | 1 | 60 | MEF2C |
| 5f28_D | 1e-17 | 11 | 70 | 1 | 60 | MEF2C |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
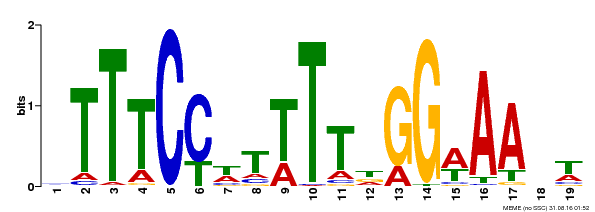
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA8623 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004516227.1 | 1e-150 | agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | A0A1S2Z743 | 1e-149 | A0A1S2Z743_CICAR; agamous-like MADS-box protein AGL15 | ||||
| STRING | XP_004516227.1 | 1e-150 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3582 | 32 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 3e-68 | AGAMOUS-like 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA8623 |



