 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp5g04800 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 354aa MW: 39701.3 Da PI: 8.878 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 127.2 | 6.3e-40 | 62 | 139 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC++dls+ak+y++rhkvC +hsk+++v+vsgl+qrfCqqCsrfh+lsefD ekrsCrrrLa hnerrrk+qa
Tp5g04800 62 RCQVEGCRMDLSNAKTYYSRHKVCCIHSKSSKVIVSGLHQRFCQQCSRFHQLSEFDLEKRSCRRRLACHNERRRKPQA 139
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.5E-31 | 56 | 124 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 30.915 | 60 | 137 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.14E-37 | 61 | 141 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.3E-29 | 63 | 136 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MELLMGSGQN RTESGASSST ESSSLCGGLR FGQKIYFEDG SGSGPGLNKN RLNTGRKSTI 60 ARCQVEGCRM DLSNAKTYYS RHKVCCIHSK SSKVIVSGLH QRFCQQCSRF HQLSEFDLEK 120 RSCRRRLACH NERRRKPQAT TTLLTSRYSR IAPSLYGNGS TAMIRSVLGD TTTWSTARSV 180 MRRSAPWQID PERESHQLMN VFSHGSSSFS TTCPEMMNNN STDSSCALSL LSNSNQNQQQ 240 QQLQASTNIW RPSSDFDSMI ADRVTMAQPP PVSIHNQYLN QTWEFMSGEK SNSHYMSSVL 300 GLSQVSEPVD LQISNGTTMG GFELSLHQQV MRQYMEPENT RAYDSSPQLF NWSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-28 | 63 | 136 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00412 | DAP | Transfer from AT3G57920 | Download |
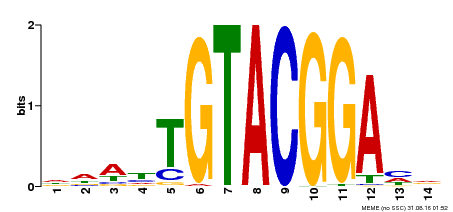
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp5g04800 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR157. {ECO:0000305|PubMed:12202040}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002878178.1 | 0.0 | squamosa promoter-binding-like protein 15 | ||||
| Swissprot | Q9M2Q6 | 0.0 | SPL15_ARATH; Squamosa promoter-binding-like protein 15 | ||||
| TrEMBL | D7LVY0 | 0.0 | D7LVY0_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.5__2235__AT3G57920.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2144 | 27 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57920.1 | 0.0 | squamosa promoter binding protein-like 15 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



