 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp5g07440 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 341aa MW: 38204 Da PI: 9.6968 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 56.1 | 9.2e-18 | 115 | 164 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta+ Aa+a+++a+ k++g
Tp5g07440 115 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAYAAARAYDRAAIKFRG 164
78*******************......55************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.9E-9 | 115 | 164 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.35E-15 | 115 | 172 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 17.332 | 116 | 172 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.7E-16 | 116 | 171 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.7E-30 | 116 | 178 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.96E-11 | 116 | 171 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MLDLSLGIFS NYQEDQDRKV PLISNAGEEE SNLSSSSSSS TDHGKAFITF GILKRHDDVV 60 PPPPKETGDL FPVADARRNI EFSVDDSHWL NLSSLQRNKQ QMVKKSRRGP RSRSSQYRGV 120 TFYRRTGRWE SHIWDCGKQV YLGGFDTAYA AARAYDRAAI KFRGLDADIN FVVDDYRQDI 180 DKMKNLNKVE FVQTLRRESA SFGRGSSKYK GLTLQKCTQF KTHHDQIHLF QNRGWDAAAI 240 KYNELGKGEG VMKFGPHIKG KAHNDLELSL GLSPSSENIK LTTSDYYKGS NLSTMGFLGK 300 QSSIYLPVTT MIPLKTVAAS SGFPFITMTT SSSSMSACFD P |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Repressor of flowering. {ECO:0000250, ECO:0000269|PubMed:14573523}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00075 | ChIP-chip | Transfer from AT3G54990 | Download |
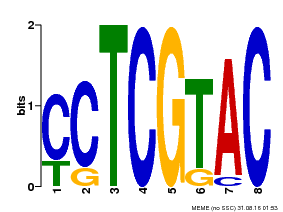
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp5g07440 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172 after photoperiod changement. {ECO:0000269|PubMed:14573523}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY576275 | 0.0 | AY576275.1 Arabidopsis thaliana AP2 domain transcription factor (SMZ) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010427236.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor SMZ | ||||
| Swissprot | Q6PV68 | 1e-167 | SMZ_ARATH; AP2-like ethylene-responsive transcription factor SMZ | ||||
| TrEMBL | A0A1J3JSM5 | 0.0 | A0A1J3JSM5_NOCCA; AP2-like ethylene-responsive transcription factor SMZ (Fragment) | ||||
| STRING | XP_010427236.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10858 | 18 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G54990.1 | 1e-165 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



