 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp5g11150 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 371aa MW: 38762.5 Da PI: 7.063 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 231.5 | 1.5e-71 | 136 | 286 | 4 | 153 |
DUF702 4 gtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa.aeaaskrkrelkskkqsalsstklssaeskkelet 102
+++sCqdCGnqakkdCah RCRtCCksrgf+C thv+stWvpaakrrerqqql++ + ++++++ ++ +kr+re+ +++s+l +t+++++++ + le+
Tp5g11150 136 NGVSCQDCGNQAKKDCAHIRCRTCCKSRGFECPTHVRSTWVPAAKRRERQQQLTTVQPQTQQRRgDSGVPKRQRENLPATSSSLVCTRIPTHHNASALEI 235
4679*************************************************99988777766578899****************************** PP
DUF702 103 sslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGle 153
++P evss+avfrcvrvssv+dgeee+aYqtavsigGh+fkGiLyd+G+
Tp5g11150 236 GDFPGEVSSPAVFRCVRVSSVEDGEEEFAYQTAVSIGGHIFKGILYDHGPG 286
*************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 3.4E-65 | 137 | 285 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 1.1E-26 | 139 | 181 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 1.8E-27 | 238 | 285 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 371 aa Download sequence Send to blast |
MAGFFSLDGG GGGGGGGNSQ EEHRTNTNPP PPVSEAWLWY RNPNVNANTN TNAPSSSNAA 60 LGTLELWQNH NQQDIMFQHQ QHQQRLDLYS SAAGLGVGPS NHSQFDISGE TSNAGAGREA 120 ALMMMRSGGS GGGGGNGVSC QDCGNQAKKD CAHIRCRTCC KSRGFECPTH VRSTWVPAAK 180 RRERQQQLTT VQPQTQQRRG DSGVPKRQRE NLPATSSSLV CTRIPTHHNA SALEIGDFPG 240 EVSSPAVFRC VRVSSVEDGE EEFAYQTAVS IGGHIFKGIL YDHGPGSSEG GGGYNVITAG 300 ESSSGGGGAQ QLNLITAGSV TVATASSSTP NAGGICSSSA AAAVYIDPTS LYPTPINTFM 360 AGTQFFPNPR S |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). {ECO:0000269|PubMed:12361963, ECO:0000269|PubMed:16740145, ECO:0000269|PubMed:16740146, ECO:0000269|PubMed:18811619, ECO:0000269|PubMed:20154152, ECO:0000269|PubMed:22318676}. | |||||
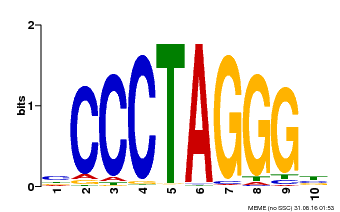
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp5g11150 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Regulated by ESR1 and ESR2. {ECO:0000269|PubMed:21976484}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU205262 | 0.0 | GU205262.1 Brassica rapa subsp. pekinensis stylish (STY1-1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013744650.1 | 0.0 | protein SHI RELATED SEQUENCE 1-like | ||||
| Swissprot | Q9SD40 | 0.0 | SRS1_ARATH; Protein SHI RELATED SEQUENCE 1 | ||||
| TrEMBL | V4NGU3 | 0.0 | V4NGU3_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006403958.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3419 | 28 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G51060.1 | 1e-143 | Lateral root primordium (LRP) protein-related | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



