 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp5g27120 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 393aa MW: 44620.1 Da PI: 7.6997 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.1 | 0.00061 | 21 | 43 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C++Cg s s k++ ++Hi++H
Tp5g27120 21 YLCQYCGISRSKKYLITSHINSH 43
56********************6 PP
| |||||||
| 2 | zf-C2H2 | 20.2 | 1.6e-06 | 67 | 89 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cg F+ + +Lk+H+++H
Tp5g27120 67 HTCQECGAEFKKPAHLKQHMQSH 89
79*******************99 PP
| |||||||
| 3 | zf-C2H2 | 16.1 | 3.1e-05 | 95 | 119 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C dC+ s++rk++L+rH+ tH
Tp5g27120 95 FACYvsDCTSSYRRKDHLNRHLLTH 119
668888*****************99 PP
| |||||||
| 4 | zf-C2H2 | 10.9 | 0.0014 | 124 | 149 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp dC+ Fs + n rH ++ H
Tp5g27120 124 FKCPmeDCKSEFSVHGNVSRHVKKfH 149
89*********************999 PP
| |||||||
| 5 | zf-C2H2 | 20.3 | 1.5e-06 | 196 | 220 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ s+L++H+ +H
Tp5g27120 196 HVCKeiGCGKAFKYASQLQKHQGSH 220
79*******************9887 PP
| |||||||
| 6 | zf-C2H2 | 20.3 | 1.5e-06 | 286 | 311 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kC+ C+ Fs snL++H+++ H
Tp5g27120 286 FKCEveGCSSKFSKASNLQKHMKVvH 311
89*********************988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 25 | 21 | 43 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-4 | 67 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 14.004 | 67 | 94 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0015 | 67 | 89 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 69 | 89 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.78E-10 | 80 | 121 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.0E-12 | 87 | 121 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.681 | 95 | 124 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.34 | 95 | 119 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 97 | 119 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 8.767 | 124 | 154 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.3 | 124 | 149 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 126 | 149 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.35E-6 | 191 | 222 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-7 | 191 | 220 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.803 | 196 | 225 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0028 | 196 | 220 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 198 | 220 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.97 | 228 | 253 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.3E-4 | 230 | 251 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 230 | 253 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 26 | 256 | 277 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 5.15E-9 | 267 | 323 | No hit | No description |
| PROSITE profile | PS50157 | 12.944 | 286 | 316 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.2E-9 | 286 | 311 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 2.1E-4 | 286 | 311 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 288 | 311 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-4 | 312 | 337 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 8.1 | 317 | 343 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 319 | 343 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 393 aa Download sequence Send to blast |
MAGEAANVDV KPSKTKDIRN YLCQYCGISR SKKYLITSHI NSHHKMEVEE ERVDEACEDE 60 EAASGKHTCQ ECGAEFKKPA HLKQHMQSHS LERPFACYVS DCTSSYRRKD HLNRHLLTHK 120 GKLFKCPMED CKSEFSVHGN VSRHVKKFHC NGDGSKDDTG NGDINKDHIN NSDSRKEDLG 180 DGDSRSSDCS TGQKQHVCKE IGCGKAFKYA SQLQKHQGSH VKLDSVEAFC SEPGCMKYFT 240 NEECLKAHIR SCHQHIDCEI CGSQHLKKNI KRHLRTHDED SSPGEFKCEV EGCSSKFSKA 300 SNLQKHMKVV HEDIRPFVCG FPGCGMRFAY KHVRNNHENS GCHMYTCGDF VEADEDFTSR 360 PRGGVKRKQV TAEMLVRKRV MPPQFDSEEH EIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wjq_D | 6e-18 | 19 | 337 | 7 | 275 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
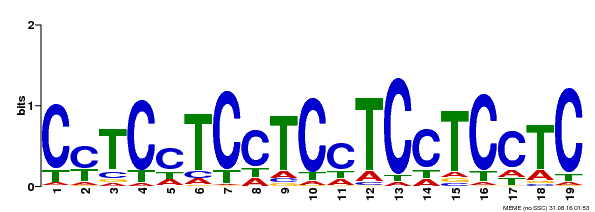
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp5g27120 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006301795.1 | 0.0 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 0.0 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | R0IDC6 | 0.0 | R0IDC6_9BRAS; Uncharacterized protein | ||||
| STRING | Cagra.4006s0023.1.p | 0.0 | (Capsella grandiflora) | ||||
| STRING | XP_006301795.1 | 0.0 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4714 | 28 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 0.0 | transcription factor IIIA | ||||



