 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp5g32010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 585aa MW: 67085.7 Da PI: 7.295 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 93.9 | 1.5e-29 | 42 | 126 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW++ e+laL+++r+em++++r++ lk+plWee+s+km+e g++r++k+Ckek+en+ k++k++keg+ ++ ++ +t+++f++lea
Tp5g32010 42 RWPRPETLALLRIRSEMDKAFRDSTLKAPLWEEISRKMMELGYKRKAKKCKEKFENVYKYHKRTKEGRTGK--PEGKTYRFFEELEA 126
8********************************************************************95..55568******985 PP
| |||||||
| 2 | trihelix | 108.8 | 3.6e-34 | 407 | 492 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+k ev aLi++r+++e +++++ k+plWee+s+ mr+ g++rs+k+Ckekwen+nk++kk+ke++kkr + +s+tcpyf+qlea
Tp5g32010 407 RWPKTEVEALIRIRKNLEANYQENGTKGPLWEEISAGMRRLGYNRSAKRCKEKWENINKYFKKVKESNKKR-PLDSKTCPYFHQLEA 492
8*********************************************************************8.9************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.61 | 39 | 101 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 6.69 | 41 | 99 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 4.47E-16 | 41 | 106 | No hit | No description |
| Pfam | PF13837 | 5.0E-20 | 41 | 126 | No hit | No description |
| PROSITE profile | PS50090 | 7.526 | 400 | 464 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 9.2E-4 | 404 | 466 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-4 | 406 | 463 | IPR009057 | Homeodomain-like |
| CDD | cd12203 | 2.25E-26 | 406 | 471 | No hit | No description |
| Pfam | PF13837 | 7.3E-24 | 406 | 493 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 585 aa Download sequence Send to blast |
MSGNSSGLLD SSGGGGVGGS GEEEKDMKME ETGEVGGGGG NRWPRPETLA LLRIRSEMDK 60 AFRDSTLKAP LWEEISRKMM ELGYKRKAKK CKEKFENVYK YHKRTKEGRT GKPEGKTYRF 120 FEELEAFETL NSYPPEPESQ PAKSPAATMT TTASATTSLI PWISSNNPST EKNSLPSKHH 180 HHHHQFSVKP IATSPTFLAK QPSSTTPFPF YSNNHTTTVD TGFKPTSNDL INNVSSLNLF 240 SSSTSSSTAS DEEEDHHQEK RSRKKRKYWK GLFTKLTKEL MEKQEKMQKR FLETLENRER 300 ERISREEAWR VQEVARINRE HEILVHERSN AADKDTAIIS FLQKISGGQQ QPRQQPQQNH 360 KVAQRKQYQS EHSITFESKE PRPVLLDTTM KMGNYDGNHS VSPSSSRWPK TEVEALIRIR 420 KNLEANYQEN GTKGPLWEEI SAGMRRLGYN RSAKRCKEKW ENINKYFKKV KESNKKRPLD 480 SKTCPYFHQL EALYNERNKS GALPILPLMV TPQKQLLLPQ ETQTEFETDQ QDKVVNKEVE 540 EEGESEEDDY EDDEEEGEGD NETSEFEIVL NKTSSPMDIN NNLFT |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 84 | 92 | KRKAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
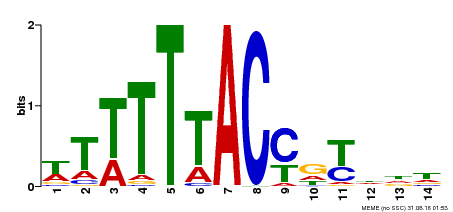
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp5g32010 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY087117 | 0.0 | AY087117.1 Arabidopsis thaliana clone 3190 mRNA, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_177815.1 | 0.0 | Duplicated homeodomain-like superfamily protein | ||||
| Swissprot | Q39117 | 0.0 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | A0A087HJ00 | 0.0 | A0A087HJ00_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087HJ00 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5995 | 28 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 0.0 | Trihelix family protein | ||||



