 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp6g20540 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 433aa MW: 49007.6 Da PI: 8.4697 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.5 | 2.8e-17 | 105 | 151 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ed +l+++v+q+G + W+ I++++ gR +kqc++rw+++l
Tp6g20540 105 KGQWTADEDRKLIRLVRQHGERKWAMISEKLE-GRAGKQCRERWHNHL 151
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.3 | 6.3e-17 | 158 | 199 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ W++eE+ lv+a+++ G++ W+ Ia+ ++ gRt++++k++w+
Tp6g20540 158 DGWSEEEERVLVEAHTRIGNK-WAEIAKLIP-GRTENSIKNHWN 199
56*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 28.634 | 100 | 155 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.31E-29 | 102 | 198 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-15 | 104 | 153 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-26 | 106 | 158 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.53E-15 | 108 | 151 | No hit | No description |
| Pfam | PF13921 | 1.8E-18 | 108 | 166 | No hit | No description |
| SMART | SM00717 | 3.7E-14 | 156 | 204 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.493 | 156 | 206 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-20 | 159 | 204 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.54E-11 | 160 | 202 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 433 aa Download sequence Send to blast |
MEHRRLVHGA APPLTAVERF LYGQKNDALC SKKQECSRDR HSPIVKKTAA IEIRNDNKEN 60 MTFGPRKEKH LVVHGRSSIG DMVVKDATRD YQNPTKKRPY KNLIKGQWTA DEDRKLIRLV 120 RQHGERKWAM ISEKLEGRAG KQCRERWHNH LRPDIKKDGW SEEEERVLVE AHTRIGNKWA 180 EIAKLIPGRT ENSIKNHWNA TKRRQNSKRK HKRQVNADNN DSDLSPAAKR PCILQDYIKS 240 IDNNVNNKDN DDNKNENSIS VLSTPNLDQI YSDGDSASSV LGDPYDEELV YLQNIFANHP 300 ISLENIGLSQ TSDEVNQSSS SGFMIKNPHP NSHNTVGAHH HDQAMFAVPA NTPSPHLPSD 360 IYLSCLLNGT TSSYSNPHFP SYSSSTSSTT VEQEGHSELL VPQANFASER REMDLIEMLA 420 GSIQGSNVSF PLF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 3e-44 | 103 | 205 | 2 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 1e-43 | 103 | 205 | 56 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-43 | 103 | 205 | 56 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 3e-44 | 103 | 205 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-44 | 103 | 205 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 201 | 212 | KRRQNSKRKHKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for female gametophyte fertility. Acts redundantly with MYB64 to initiate the FG5 transition during female gametophyte development. The FG5 transition represents the switch between free nuclear divisions and cellularization-differentiation in female gametophyte, and occurs during developmental stage FG5. {ECO:0000269|PubMed:24068955}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00567 | DAP | Transfer from AT5G58850 | Download |
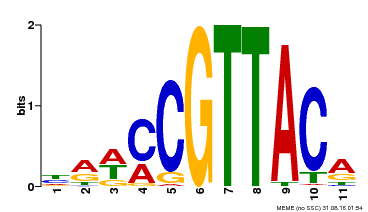
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp6g20540 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006401032.1 | 0.0 | transcription factor MYB119 | ||||
| Swissprot | Q9FIM4 | 0.0 | MY119_ARATH; Transcription factor MYB119 | ||||
| TrEMBL | V4KWZ1 | 0.0 | V4KWZ1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006401032.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4265 | 26 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58850.1 | 0.0 | myb domain protein 119 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



