 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp6g21770 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 495aa MW: 54123.2 Da PI: 7.697 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.2 | 7.6e-17 | 167 | 216 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Tp6g21770 167 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAVKFRG 216
78*******************......55***********************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.2E-8 | 167 | 216 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.61E-11 | 167 | 223 | No hit | No description |
| SuperFamily | SSF54171 | 2.42E-15 | 167 | 224 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.3E-30 | 168 | 230 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.359 | 168 | 224 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-16 | 168 | 224 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 495 aa Download sequence Send to blast |
MLDLNLDVES AESTQNRRDS AAVKGVSGAI LNQMDESVTS NSSVVNAEAS SCIDGEEELC 60 STRVVKFQFE ILKGGAGGEE EEEEQEDRSG EMMTKEFFPV AKGEGDGLNF LDRSAQGSRS 120 SVDISFQRGK QGGDFLGDGS GGDAARVMQP PAQPVKKSRR GPRSKSSQYR GVTFYRRTGR 180 WESHIWDCGK QVYLGGFDTA HAAARAYDRA AVKFRGLEAD INFIISDYEE DLKQMANLSK 240 EEVVQVLRRQ SSGFSRNNSR YQGVALQKIG GWGAQMEQLN GNMAYDKAAT KWNGREAASL 300 IEPHASRMIP EAANVKLDLN LGISLSLGDG PKQKDRALRL HHAPNSIICG RNSKMENHMA 360 AATCDTPFNF LKRSSDHLIN RHALPSAFFS PMERTPEKGF SSRSPQSFPA RTRQAQEQFS 420 GGTATTAIAT PLYSNAASSG FSLSATRPPS SSTGTLHPSQ PFLNLNMPGL YVIHPSDYAS 480 QQHHHHLMNR SQPPP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00616 | PBM | Transfer from AT5G60120 | Download |
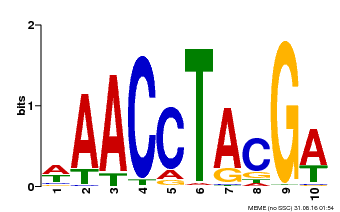
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp6g21770 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352946 | 0.0 | AK352946.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-19-B14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006400879.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE2 | ||||
| Swissprot | Q9LVG2 | 0.0 | TOE2_ARATH; AP2-like ethylene-responsive transcription factor TOE2 | ||||
| TrEMBL | E4MWP9 | 0.0 | E4MWP9_EUTHA; mRNA, clone: RTFL01-19-B14 | ||||
| TrEMBL | V4LIS4 | 0.0 | V4LIS4_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006400879.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM12836 | 17 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G60120.1 | 0.0 | target of early activation tagged (EAT) 2 | ||||



