 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp6g37110 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 273aa MW: 30999.9 Da PI: 7.6283 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 25.5 | 3.1e-08 | 29 | 74 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
+WT+eE +++ +a + + + +W ++a++++ g+t ++ + k
Tp6g37110 29 SWTQEENKKFERALAIYADDtpdRWFKVAAMIP-GKTISDVMRQYSK 74
7*****************99*************.******9998877 PP
| |||||||
| 2 | Myb_DNA-binding | 40 | 9.1e-13 | 133 | 177 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + ++G+g+W+ I+r + +t+ q+ s+ qky
Tp6g37110 133 PWTEEEHRRFLLGLLKYGKGDWRNISRNFVGSKTPTQVASHAQKY 177
8****************************999************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-4 | 25 | 73 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51293 | 7.752 | 25 | 78 | IPR017884 | SANT domain |
| SMART | SM00717 | 6.0E-8 | 26 | 78 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 5.53E-11 | 28 | 82 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.29E-7 | 29 | 75 | No hit | No description |
| Pfam | PF00249 | 5.1E-7 | 29 | 74 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.108 | 126 | 182 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.08E-15 | 128 | 181 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.9E-11 | 130 | 180 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-11 | 130 | 182 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.4E-16 | 130 | 181 | IPR006447 | Myb domain, plants |
| CDD | cd00167 | 7.01E-10 | 133 | 177 | No hit | No description |
| Pfam | PF00249 | 3.9E-10 | 133 | 177 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
METLHPLVLS HVPTSDHRFV VQEMMQSSSW TQEENKKFER ALAIYADDTP DRWFKVAAMI 60 PGKTISDVMR QYSKLEEDLF DIEAGLVPIP GYPSATLCGF DQLVSPRDFD LYRKRPNGGG 120 AREFDQDRRK GVPWTEEEHR RFLLGLLKYG KGDWRNISRN FVGSKTPTQV ASHAQKYYQR 180 QLSGAKDKRR PSIHDITTVN LLNNNLSRPS SDHDCFLQKP AEPKLGFTDR DNAAEGVMFL 240 GQSLSSVLSP YDPAFKFSAE NVYGDGGFCI TRS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-15 | 30 | 92 | 11 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
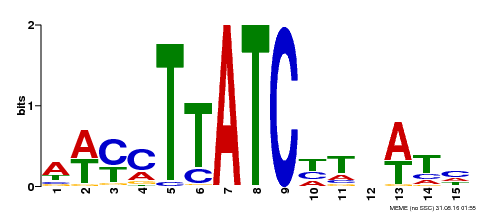
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00489 | DAP | Transfer from AT5G05790 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp6g37110 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519531 | 0.0 | AY519531.1 Arabidopsis thaliana MYB transcription factor (At5g05790) mRNA, complete cds. | |||
| GenBank | BT026054 | 0.0 | BT026054.1 Arabidopsis thaliana At5g05790 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018445578.1 | 1e-179 | PREDICTED: transcription factor DIVARICATA-like | ||||
| Swissprot | Q8S9H7 | 1e-73 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | Q9FFJ9 | 1e-177 | Q9FFJ9_ARATH; At5g05790 | ||||
| STRING | AT5G05790.1 | 1e-178 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4340 | 26 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G05790.1 | 1e-176 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



