 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_1BL_8709BB152.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 745aa MW: 82240.8 Da PI: 7.4273 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 112.1 | 3.3e-35 | 55 | 130 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+Cqv+gCead+ e+k yh+rh+vC +++a++v+++g ++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Traes_1BL_8709BB152.2 55 RCQVPGCEADIRELKGYHKRHRVCLRCAHASAVMIDGAQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 130
6************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.9E-27 | 48 | 117 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.445 | 53 | 130 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.96E-33 | 54 | 133 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.4E-27 | 56 | 130 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 745 aa Download sequence Send to blast |
MAAAAEVEEV AAEVLAGPRK KPRAGSRGSS GAVVKIGGAG GGSGVAGRGG AAEMRCQVPG 60 CEADIRELKG YHKRHRVCLR CAHASAVMID GAQKRYCQQC GKFHILLDFD EDKRSCRRKL 120 ERHNKRRRRK PDSKGTLEKE IDEQLDLSAD GSGSGEIREE NIDGASCDML ETVLSNKVLD 180 RETPVGSEDA LSSPTCTQLS LQNDRSKSIV TFAASAEACL GAKQENAKLT TSPVHDSRSA 240 YSSSCPTGRI SFKLYDWNPA EFPRRLRNQI FEWLSSMPVE LEGYIRPGCT ILTVFIAMPQ 300 HMWDKLSEDT ANLVRNLVNA PSSLLLGKGA FFVHVNNTIF QVLKDGATLM STRLEVQAPR 360 IHCVHPTWFE AGKPIELLLC GSSLDQPKFR SLLSFDGEYL KHDCCRLTSR EIFGCVKNGA 420 PTFDSHHEVF RINITQTKPD THGPGFVEVE NVFGLSNFVP ILFGSKQLCF ELERIQDVLC 480 GSSKYKSTNG EFPGITSDRC EHWKLQQTAM SGFLIEIGWL IKKPSPDEFK NLLSKTNIKR 540 WICLLEFLIQ NNFINVLEMI VKSSDNIIGS EILSNLESGR LEDHVTAFLG YVRHARTIVD 600 QRAKHNEETQ LQTRWCGDSV SDQPSLGTSV PLGKENVAAS DDFCLPSTNA ECEAEESVPL 660 VTNEAVSHRQ CRAPEMNARW LNPALVAPFP SGMMRTRLVA TVAVAAILCF TACVALFHPG 720 RVGVLAAPVK RYLFSDCPPW HHSGC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 2e-38 | 55 | 137 | 5 | 87 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 112 | 130 | KRSCRRKLERHNKRRRRKP |
| 2 | 116 | 128 | RRKLERHNKRRRR |
| 3 | 123 | 130 | NKRRRRKP |
| 4 | 124 | 129 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
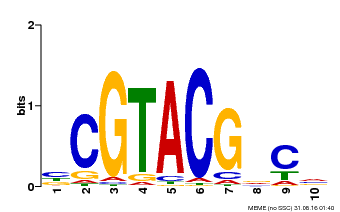
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK250312 | 0.0 | AK250312.1 Hordeum vulgare subsp. vulgare cDNA clone: FLbaf72c21, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020161545.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A446K185 | 0.0 | A0A446K185_TRITD; Uncharacterized protein | ||||
| STRING | Traes_1BL_8709BB152.2 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-136 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



