 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_1BS_403DBC53C.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 441aa MW: 48848.9 Da PI: 6.5069 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 61.6 | 1.7e-19 | 50 | 96 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT eEd++l +av+ + g++Wk+Ia++++ Rt+ qc +rwqk+l
Traes_1BS_403DBC53C.1 50 KGNWTLEEDDILRKAVEIHNGKNWKKIAECFP-DRTDVQCLHRWQKVL 96
79******************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 58.8 | 1.2e-18 | 102 | 148 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd+ +++ vk++G+ W+tIa+ ++ gR +kqc++rw+++l
Traes_1BS_403DBC53C.1 102 KGPWSKEEDDVIIQMVKKYGPTKWSTIAQALP-GRIGKQCRERWHNHL 148
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 54.8 | 2.2e-17 | 154 | 196 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+++WT+eE+ +l++a++ +G++ W+ ++ ++ gRt++ +k++w+
Traes_1BS_403DBC53C.1 154 KDAWTQEEEIRLIQAHHIYGNK-WAELSKFLP-GRTDNAIKNHWH 196
689*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.034 | 45 | 96 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.9E-17 | 49 | 98 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.9E-17 | 50 | 96 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.52E-16 | 51 | 106 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.4E-24 | 52 | 112 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.87E-15 | 52 | 96 | No hit | No description |
| PROSITE profile | PS51294 | 31.793 | 97 | 152 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.27E-32 | 99 | 195 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.8E-17 | 101 | 150 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.82E-15 | 104 | 148 | No hit | No description |
| Pfam | PF13921 | 9.0E-19 | 105 | 165 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.8E-27 | 113 | 156 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.0E-15 | 153 | 201 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 20.671 | 153 | 203 | IPR017930 | Myb domain |
| CDD | cd00167 | 1.74E-12 | 156 | 196 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-21 | 157 | 202 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 441 aa Download sequence Send to blast |
MTSDKGRTSK KAAEGSPSAL AVPDGRASSG PQKGRLSNGR TTGPARRSTK GNWTLEEDDI 60 LRKAVEIHNG KNWKKIAECF PDRTDVQCLH RWQKVLNPEL VKGPWSKEED DVIIQMVKKY 120 GPTKWSTIAQ ALPGRIGKQC RERWHNHLNP GINKDAWTQE EEIRLIQAHH IYGNKWAELS 180 KFLPGRTDNA IKNHWHSSVK KKYESYRAEG LLAQFQGLPA VVCPTGSLNV DSSSAMMQQN 240 SDGSGLNVNQ EVQDSSEFSQ SALANISSSQ VEQVDEPVGC HGQIHSNDGA HDSYSSCQEA 300 CHTKANDAAS TSPETRHQLS ISENDPDIHW QQDLSVADFD LQSELWQDIS YQSLLTAPDS 360 INAVSFFSPN YQHSACSSEV ANNFEERLYP LHTYSSSRTG TVHHQSSATS VPPSFLCSGR 420 ASQIVSSELP IVPESKEQQM T |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 2e-68 | 50 | 202 | 6 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-68 | 50 | 202 | 6 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
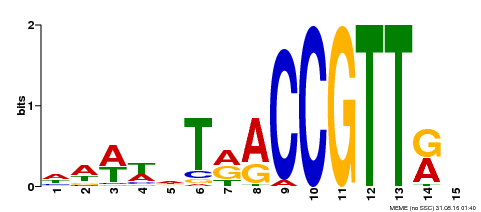
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JF951920 | 0.0 | JF951920.1 Triticum aestivum clone TaMYB35 R1R2R3-MYB protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020174664.1 | 0.0 | transcription factor MYB3R-1-like | ||||
| TrEMBL | A0A3B5YUI9 | 0.0 | A0A3B5YUI9_WHEAT; Uncharacterized protein | ||||
| STRING | Traes_1BS_403DBC53C.1 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2495 | 38 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.2 | 1e-108 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



