 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_1DS_34596D06E.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 232aa MW: 25163.6 Da PI: 6.0535 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 64.5 | 2.3e-20 | 49 | 117 | 1 | 68 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkm.rergferspkqCkekwenlnkrykkikege 68
+W++ e+ a +++r ++++++ ++k++k+lWe+vs+++ +++gf r+p qCk+kw+nl +r+k ++ +
Traes_1DS_34596D06E.1 49 QWSHAETAAFLAIRADLDHSFLSTKRNKALWEAVSARLnAQGGFARTPDQCKSKWKNLVTRFKGTASDA 117
7*************************************88999********************998765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd12203 | 7.31E-22 | 48 | 113 | No hit | No description |
| Pfam | PF13837 | 1.4E-16 | 49 | 142 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 7.3E-4 | 49 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 7.457 | 50 | 107 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 232 aa Download sequence Send to blast |
MDPFHHLNTA FSNPYHPLLS SSPPHPYHPP PPLPPPPAAD PPERERLPQW SHAETAAFLA 60 IRADLDHSFL STKRNKALWE AVSARLNAQG GFARTPDQCK SKWKNLVTRF KGTASDAQPE 120 GGDHPAGAAA RGSFPFHDEM RRIFDARVER ARALEAKKAK GKDAAARRDP DDDGGGEGDE 180 GEDDEEEAEM VDEEEGRADA EARGAGRKRR RKAAPARTAS AGGGVEIGEV EA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 206 | 210 | RKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00481 | DAP | Transfer from AT5G01380 | Download |
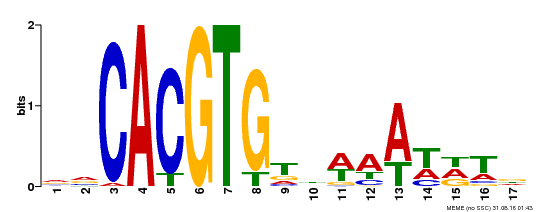
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375948 | 0.0 | AK375948.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3110L24. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020178214.1 | 1e-166 | trihelix transcription factor GT-3b-like | ||||
| TrEMBL | A0A3B5ZPU7 | 1e-164 | A0A3B5ZPU7_WHEAT; Uncharacterized protein | ||||
| STRING | Traes_1DS_34596D06E.1 | 1e-165 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12595 | 31 | 34 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G01380.1 | 1e-28 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



