 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_1DS_BB8508CC6.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 235aa MW: 24674.8 Da PI: 8.4898 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 125.8 | 2e-39 | 56 | 155 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqle 88
+CaaCk+lrr+Ca++C+++p f +p+kfa vhk+FGasnv+k+l +++e++r da++slvyeA+ r+rdPvyG++g il+lqqq++
Traes_1DS_BB8508CC6.1 56 PCAACKLLRRRCAQECPFSPFFSPLEPHKFASVHKVFGASNVSKMLLEVHESQRGDAANSLVYEANLRLRDPVYGCMGAILTLQQQVH 143
7*************************************************************************************** PP
DUF260 89 qlkaelallkee 100
l+aela++++e
Traes_1DS_BB8508CC6.1 144 ALEAELAAVRAE 155
********9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 25.114 | 55 | 156 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 2.2E-39 | 56 | 153 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 235 aa Download sequence Send to blast |
MSTASEWQQD HEVGKKIKSE SAAEADRMMA AARRSSSLPA AGVGPGSTPS FNTMTPCAAC 60 KLLRRRCAQE CPFSPFFSPL EPHKFASVHK VFGASNVSKM LLEVHESQRG DAANSLVYEA 120 NLRLRDPVYG CMGAILTLQQ QVHALEAELA AVRAEILKHR YRPAAAASAV PNVPPSSHAS 180 QLLAAGGHRP AGSLGLAAPG AGPVPSASSS TTVYATASSS TDYSSITHEN VPYFG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 3e-40 | 56 | 159 | 11 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 3e-40 | 56 | 159 | 11 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00283 | DAP | Transfer from AT2G30340 | Download |
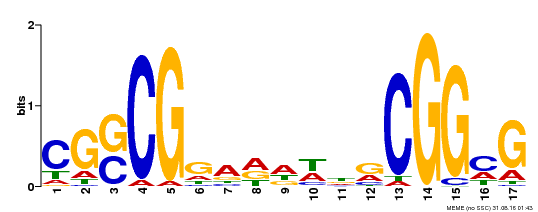
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP095831 | 1e-117 | FP095831.1 Phyllostachys edulis cDNA clone: bphyst036h20, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020167556.1 | 1e-172 | LOB domain-containing protein 15-like | ||||
| Swissprot | Q9AT61 | 3e-69 | LBD13_ARATH; LOB domain-containing protein 13 | ||||
| TrEMBL | A0A3B5ZSF7 | 1e-171 | A0A3B5ZSF7_WHEAT; Uncharacterized protein | ||||
| STRING | Traes_1DS_BB8508CC6.1 | 1e-172 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2630 | 38 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G30340.1 | 3e-56 | LOB domain-containing protein 13 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



