 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_2AS_4C4190DEB.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 917aa MW: 103030 Da PI: 9.0968 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 166.2 | 5.3e-52 | 25 | 139 | 4 | 117 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahsee 90
e +rw +++ei+a+L+n+e++++++++ ++p sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa++e+
Traes_2AS_4C4190DEB.2 25 EaASRWFRPNEIYAVLANHERFKVHAQPIDKPVSGTIVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEERVHVYYARGED 112
4489************************************************************************************ PP
CG-1 91 nptfqrrcywlLeeelekivlvhylev 117
np+f rrcywlL++e e+ivlvhy+++
Traes_2AS_4C4190DEB.2 113 NPNFFRRCYWLLDKEAERIVLVHYRQT 139
************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 73.93 | 19 | 145 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.8E-62 | 22 | 140 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.1E-44 | 26 | 138 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.23E-10 | 331 | 418 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.10E-16 | 514 | 624 | No hit | No description |
| SuperFamily | SSF48403 | 1.23E-17 | 523 | 630 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.5E-18 | 526 | 629 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.969 | 532 | 636 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.1E-8 | 538 | 628 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 3.6E-6 | 565 | 594 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.915 | 565 | 597 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 70 | 604 | 633 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 6.9E-9 | 716 | 782 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 6.833 | 716 | 742 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.18 | 731 | 753 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.627 | 732 | 758 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0019 | 734 | 753 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0095 | 754 | 776 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.493 | 755 | 779 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0051 | 756 | 776 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.41 | 835 | 857 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.608 | 837 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 917 aa Download sequence Send to blast |
DPLLRSEIHG FITYADLNFE KLKAEAASRW FRPNEIYAVL ANHERFKVHA QPIDKPVSGT 60 IVLYDRKVVR NFRKDGHNWK KKKDGKTVQE AHEKLKIGNE ERVHVYYARG EDNPNFFRRC 120 YWLLDKEAER IVLVHYRQTS EENAIAHPST EAEAEAPTMN VIQYYTSPIS ANSASVHTEI 180 SFSPPAPEEI NSHGGSAISS DTGGSSLEEF WVHLLESSMT KDTACGASVA FSQQIKCGTK 240 DSGNDTDSTN NVHVNHAGAL EHQVDQSQYP LTSDLDSQSH QFATSLRKTP VDGDIPNDVP 300 ARENSLGLWK YLDDDSPCLG DNIVSNGKIF NITDFSPEWA CSTEHTKILV IGDYYEQYKH 360 LAGSNIYGIF GDNCVAANMV QTGVYRFMVG PHTAGRVDFY LTLDGKTPIS EVLNFEYRSV 420 PGSSLHSELK PLEDEYTKSK LQMQMRLARL LFVTNKKKIA PKLLVEGSKV SNLILASPEK 480 EWMDLWKIAG DSEGTSVHAT EDLLELVLRN RLQEWLLERV IGGHKSTGRD DLGQGPIHLC 540 SFLGYTWAIR LFSVSGFSLD FRDSSGWTAL HWAAYHGREK MVAALLSAGA NPSLVTDPTA 600 VSPGGCTPAD LAARQGYVGL AAYLAEKGLT AHFESMSLSK GTKQSPSRTK LTKVHSEKFE 660 NLTEQELCLK ESLAAYRNAA DAASNIQAAL RDRTLKLQTK AILLANPEMQ ATAIVAAMRI 720 QHAFRNYNRK KEMRAAARIQ NHFRTWKVRR NFKNMRRQAI RIQAAYRGHQ VRRQYRKVIW 780 SVGVVEKAIL RWRKKRKGLR GIANGMPAEM TVDVEAASTA EEGFFQASRQ QAEDRFNRSV 840 VRVQALFRCH RAQHEYRRMR IAHEEAKLEF SKGQQQAPAC RRTILLILLA ESGVHVDAGT 900 DHVVKANNKC YLGQRPN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
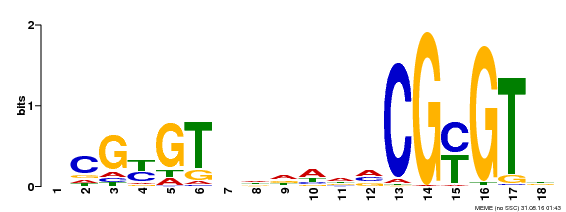
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375447 | 0.0 | AK375447.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3094H17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020186933.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A3B6AWM5 | 0.0 | A0A3B6AWM5_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446KWY3 | 0.0 | A0A446KWY3_TRITD; Uncharacterized protein | ||||
| STRING | Traes_2AS_4C4190DEB.2 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



