 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_4BL_941E7DE4D.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 684aa MW: 73606.6 Da PI: 6.617 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 89.9 | 2.2e-28 | 203 | 256 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av++L G++kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
Traes_4BL_941E7DE4D.2 203 KPRVVWSVELHQQFVNAVNHL-GIDKAVPKKILELMNVPGLTRENVASHLQKFRL 256
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 76.7 | 8.5e-26 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeeda 86
vl+vdD+++ + +l+++l + +y +++++++++ al++l+e++ +D+i+ D+ mp+m+G++ll+ e++lp+i+++a + +++
Traes_4BL_941E7DE4D.2 19 VLVVDDDQTCLAVLKRMLVQCRY-DATTCSQATRALAMLRENRgaFDVIISDVHMPDMNGFRLLELVGL-EMDLPVIMMSADSRTDLV 104
89*********************.*****************9999*******************87754.558*************** PP
HHHHHTTESEEEESS--HHHHHH CS
Response_reg 87 lealkaGakdflsKpfdpeelvk 109
+ +k Ga d+l Kp+ +eel +
Traes_4BL_941E7DE4D.2 105 MKGIKHGACDYLIKPVRMEELKN 127
********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 1.3E-181 | 2 | 669 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 4.2E-41 | 16 | 159 | No hit | No description |
| SuperFamily | SSF52172 | 6.97E-35 | 16 | 137 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 4.4E-30 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.328 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 9.3E-23 | 19 | 127 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 6.10E-27 | 20 | 132 | No hit | No description |
| PROSITE profile | PS51294 | 11.948 | 200 | 259 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.3E-30 | 200 | 261 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.36E-19 | 201 | 260 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.0E-25 | 203 | 256 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.8E-7 | 205 | 254 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 684 aa Download sequence Send to blast |
MAPPEEAGGG DQFPVGMKVL VVDDDQTCLA VLKRMLVQCR YDATTCSQAT RALAMLRENR 60 GAFDVIISDV HMPDMNGFRL LELVGLEMDL PVIMMSADSR TDLVMKGIKH GACDYLIKPV 120 RMEELKNIWQ HVVRKKFGGN KEHEHSGSLD DTDRNRLTNN DNEYASSAND GAEDSWKSQK 180 KKRDKEEDDS ELESGDPSNN SKKPRVVWSV ELHQQFVNAV NHLGIDKAVP KKILELMNVP 240 GLTRENVASH LQKFRLYLKR IAQHHAGMTN PYGVPASSAQ VASLGGLDFQ ALAASGQIPP 300 QALAALQDEL LGRPTNSLVL PGRDQSSLRL AAVKGNKPHG EQIAFGQPIY KVQNNSYAAL 360 PQNSSAVGRM PSFSAWPNNK LGMSDSMSAL GNVNNSQNSN MVLHELQQQP DTMLSGTLHT 420 LDVKPSGIVM PSQSLNTFPA SEGLSPNQNS LIIPSQSSGF LTGIPPSMKP ELVLPTSQSS 480 NSLLGGIDLI NQASTSQPFI SSHGGNLPGL MNRNSNVMSS QGISSFQTGN TPYLVNQNPM 540 GVGSKPPGVL KTESTDSLNQ SYAYANHIDS GLLSSQSKNA QFGFLQSPND VTGGWSSFQN 600 MDGYRNTVGP SQPVSSSSSF QSSNAALGKL PDQGRGKNLG FVGKGTCIPN RFAVDEIESP 660 TNSLSHSIGS SGDIPDMFGF SGQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-23 | 202 | 262 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
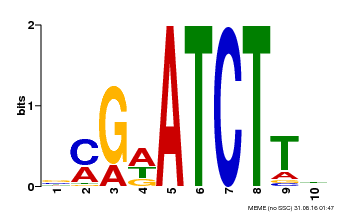
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JF951927 | 0.0 | JF951927.1 Triticum aestivum clone TaMYB44 MYB-related protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003558456.1 | 0.0 | two-component response regulator ORR21 | ||||
| Swissprot | A2XE31 | 0.0 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A3B6IV82 | 0.0 | A0A3B6IV82_WHEAT; Two-component response regulator | ||||
| TrEMBL | A0A446SAN4 | 0.0 | A0A446SAN4_TRITD; Two-component response regulator | ||||
| STRING | Traes_4BL_941E7DE4D.2 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5796 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-108 | response regulator 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



