 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_4BL_A37F6FD08.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 990aa MW: 110355 Da PI: 6.0376 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 141.2 | 3.1e-44 | 2 | 86 | 34 | 118 |
CG-1 34 pksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
p sgsl+L++rk++r+frkDG++w+kkkdgktv+E+he+LK g+++vl+cyYah+een +fqrr+yw+Lee++ +ivlvhylevk
Traes_4BL_A37F6FD08.3 2 PASGSLFLFDRKVLRFFRKDGHNWRKKKDGKTVKEAHERLKSGSIDVLHCYYAHGEENINFQRRSYWMLEEDYMHIVLVHYLEVK 86
89********************************************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01076 | 3.3E-44 | 1 | 86 | IPR005559 | CG-1 DNA-binding domain |
| PROSITE profile | PS51437 | 64.582 | 1 | 91 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.4E-37 | 2 | 85 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF01833 | 6.5E-8 | 396 | 481 | IPR002909 | IPT domain |
| CDD | cd00102 | 5.52E-5 | 396 | 483 | No hit | No description |
| Gene3D | G3DSA:2.60.40.10 | 8.2E-7 | 396 | 483 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 8.71E-18 | 397 | 482 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.77E-13 | 571 | 692 | No hit | No description |
| SuperFamily | SSF48403 | 1.09E-16 | 591 | 695 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.5E-17 | 594 | 697 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.21 | 600 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.736 | 633 | 665 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.013 | 633 | 662 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1500 | 672 | 701 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.6E-8 | 803 | 857 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.036 | 806 | 828 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.498 | 807 | 836 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0032 | 808 | 827 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 829 | 851 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 830 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.9E-4 | 832 | 851 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 990 aa Download sequence Send to blast |
MPASGSLFLF DRKVLRFFRK DGHNWRKKKD GKTVKEAHER LKSGSIDVLH CYYAHGEENI 60 NFQRRSYWML EEDYMHIVLV HYLEVKAGKS SSRTRGHDNM LQGAYVDSPL SHLPSQSTDG 120 ESSLSGRASE YEAESADIYS GGAGYHSISR MQQHENGGGS IIDASVVSSY SPASSVGNHQ 180 GLRATSPNTG FYSHYQDNSP VIHNESTLGI TFNGPSTQFD LSSWNEMTKL NKGIHQLPPY 240 QSHVPSEQPP FTEGPGIESF SFDEVYSNGL DIKDGGHADT DREALWQLPS ATDSTFATVD 300 SFEQNNKLLE EAINFPVLKT QSSNLSDILK DSFKKSDSFT RWMSKELAEV DDSQVKSSSG 360 LYWNSEDADN IIGASSRDQL DQFTLDPMVA QDQLFSITEY FPSWTYAGSK TRVLVTGRFL 420 TSDEVIKLKW SCMFGEVEVP ADILADGTLR CYSPSHKPGR VPFYVTCSNR LACSEVREFE 480 YRPSDSQYMD APSPHGATNK IYLQARLDEL LSLGQDEQDE FQAALSNPTK ELIDLNKKIT 540 SLMTNNDQWS ELLKFADDNQ LAPDDRQDQF VESGMKEKLH IWLLRKAGGG GKGPSVLDDE 600 GQGVLHLAAA LGYDWAIRPT ITAGVSINFR DVHGWTALHW AAFCGRERTV VALIALGAAP 660 GALTDPRPDF PSGRTPADLA SFNGHKGISG FLAEFSLTSH LQTLNLKEAM GSNASEVSGL 720 PGIGDVTGRI ASPSAGQGLQ AGSMGDSLGA VRNAAQAAAR IYQVFRVQSF QRKQAVQYED 780 DNGVISDERA MSLLSYKPSK PGQFDPMHAA ATRIQNKFRG WKGRKEFLLI RQRIVKIQAH 840 VRGHQVRKHY RKIIWSVGIV EKVILRWRRR GAGLRGFRST EGATDSSTRS SSVDVTPVKP 900 AEDDYNFLQE GRKQTEERLQ RALARVKSMV QYPEARDQYQ RILTVVTKMQ ESQPVEESML 960 EESTEMDEGF LMSEFKELWD DDMPPLPGYF |
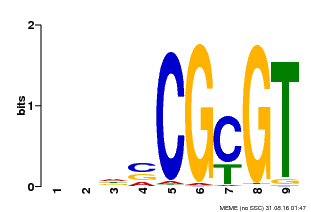
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK365210 | 0.0 | AK365210.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2032B14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020179695.1 | 0.0 | calmodulin-binding transcription activator 3-like isoform X1 | ||||
| TrEMBL | A0A3B6IU44 | 0.0 | A0A3B6IU44_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A446SF68 | 0.0 | A0A446SF68_TRITD; Uncharacterized protein | ||||
| STRING | Traes_4BL_A37F6FD08.3 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



