 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Traes_5DS_5DEA5C9E3.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 497aa MW: 52829 Da PI: 8.1601 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.1 | 1.4e-31 | 229 | 285 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgs++prsYY+Ct+++Cpvkkkve+++ d+++ ei+Y+g+Hnh+
Traes_5DS_5DEA5C9E3.1 229 DDGYNWRKYGQKVVKGSDCPRSYYKCTHPSCPVKKKVEHAE-DGQISEIIYKGKHNHQ 285
8**************************************99.***************8 PP
| |||||||
| 2 | WRKY | 108 | 4.6e-34 | 385 | 443 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+agC+v+k++er+++dpk+v++tYeg+Hnhe
Traes_5DS_5DEA5C9E3.1 385 LDDGYRWRKYGQKVVKGNPHPRSYYKCTFAGCNVRKHIERASSDPKAVITTYEGKHNHE 443
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.8E-28 | 225 | 287 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.1E-25 | 227 | 287 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-33 | 228 | 286 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.8E-24 | 229 | 285 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.192 | 229 | 287 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.5E-37 | 371 | 445 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.88E-29 | 377 | 445 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.413 | 380 | 445 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.0E-40 | 385 | 444 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.1E-26 | 386 | 443 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 497 aa Download sequence Send to blast |
MSARPPPPPR PRLSLPPRST GESLFSGTGD ASPGPLTLAS ALFPSDADAD AGGGGGGSGA 60 SSGAGPTSFT QLLIGSLSQP PPHQQQQQQQ ERGRGGVARA GPALSVAPPA GAAVFTVPPG 120 LSPSGFFDSP GLIFSPAMGG FGMSHQQALA QVTAQASHSP HRMFDHTEQP SFSAAATSSG 180 ALQNMSSAAN VAEMSEMATT ISNNEHAAFQ SAEASHRYQV PAPVDKPADD GYNWRKYGQK 240 VVKGSDCPRS YYKCTHPSCP VKKKVEHAED GQISEIIYKG KHNHQRPPNK RAKDGSSSAA 300 EQNEQSNDTA SGLSGVRRDQ EAVYGMSEQL SGLSDGDDKD DGESRPNEVD DRENDCKRRN 360 IQISSQKALT ESKIIVQTTS EVDLLDDGYR WRKYGQKVVK GNPHPRSYYK CTFAGCNVRK 420 HIERASSDPK AVITTYEGKH NHEPPVGRGS NQNGGNSNRS QQKGPNSMSS NQASHTGTDL 480 GNIKQGQIGV LQFKREE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-38 | 229 | 445 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-38 | 229 | 445 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
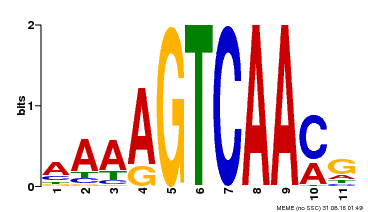
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX216840 | 0.0 | JX216840.1 Hordeum vulgare WRKY61 protein (WRKY61) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020168255.1 | 0.0 | probable WRKY transcription factor 4 | ||||
| Swissprot | Q9ZQ70 | 1e-124 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A3B6MN80 | 0.0 | A0A3B6MN80_WHEAT; Uncharacterized protein | ||||
| STRING | Traes_5DS_5DEA5C9E3.1 | 0.0 | (Triticum aestivum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1345 | 38 | 122 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03340.1 | 1e-113 | WRKY DNA-binding protein 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



