 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang0004ss00990.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 653aa MW: 71495.2 Da PI: 6.2827 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 79.3 | 4.6e-25 | 295 | 345 | 1 | 52 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQk 52
kpr++W+ eLH++Fv+av+qL G +kA+Pk+ilelm+v+gLt+e+v+SHLQ
Vang0004ss00990.1 295 KPRVVWSVELHQQFVSAVNQL-GLDKAVPKRILELMNVPGLTRENVASHLQG 345
79*******************.*****************************6 PP
| |||||||
| 2 | Response_reg | 57.5 | 7.6e-20 | 136 | 221 | 25 | 109 |
EEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 25 evaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGakdflsKpfdpeelvk 109
+++++++++ al+ll+e++ +D++l D+ mp+mdG++ll++ e +lp+i++++ ++ + + + ++ Ga d+l Kp+ +eel++
Vang0004ss00990.1 136 IITTCTEATVALNLLRERKgcFDVVLSDVHMPDMDGYKLLEHVGLEM-DLPVIMMSGDSTTSAVMKGIRHGACDYLIKPVREEELRN 221
7899**************9************************6644.8************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50110 | 25.08 | 98 | 227 | IPR001789 | Signal transduction response regulator, receiver domain |
| SMART | SM00448 | 1.6E-9 | 123 | 223 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 2.4E-16 | 136 | 222 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 2.23E-19 | 137 | 226 | No hit | No description |
| Gene3D | G3DSA:3.40.50.2300 | 2.4E-29 | 137 | 242 | No hit | No description |
| SuperFamily | SSF52172 | 6.55E-25 | 137 | 234 | IPR011006 | CheY-like superfamily |
| PROSITE profile | PS51294 | 9.962 | 292 | 351 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.2E-16 | 292 | 348 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-25 | 292 | 344 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-21 | 295 | 345 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.8E-7 | 297 | 347 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 653 aa Download sequence Send to blast |
MAQSRSAAGA CKGATAEFPA GLRSFGLDSP LVLTSQHLLR SIMIYGNYAK WFLMLTCGEA 60 YYFKYEIGGM IMDDIVSGFR DMPSLDLVRL IDCTYGEKEL LGRFFISSTR LTCKIGYFKG 120 GLYIYEWFNS CLTVAIITTC TEATVALNLL RERKGCFDVV LSDVHMPDMD GYKLLEHVGL 180 EMDLPVIMMS GDSTTSAVMK GIRHGACDYL IKPVREEELR NIWQHVVRKI WNENKEHDNS 240 GSMEDSDWNK RGNDDTEYTS SVADAAEVVK APKKRSSLKE EDIELESDDP ATSKKPRVVW 300 SVELHQQFVS AVNQLGLDKA VPKRILELMN VPGLTRENVA SHLQGWLTRL PVVDEKFRLY 360 LKRLSGVAQQ QNGMLNAVPG TIESNLSATG RFDIQALAAA GHVPPETLAA LHAELLGRPA 420 TNILSTADQA TLLQASIGLK HSHAEHAVAY GQPFVKCPSN IGKDFPPSFL TANDMSSGYG 480 AWPSSNNTMG SAVNGSASVN QNCSFGMNAV IDYSLLSQKS NNSVSASQFS GGDTKARAVL 540 YAHTMPSTCS MSSINGSIQQ LQNSTLTFGG ARPLPSPLPM SDRQDPYGLK SGDSFDQVCL 600 RNIGFIGKGT CIPNRFVQNE IESSVSDFSQ MKVNVDSTGN ALKEEPNFIN MQK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-19 | 294 | 365 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
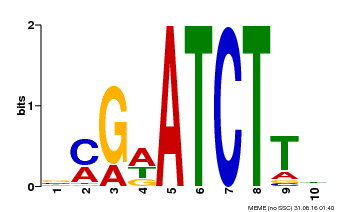
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang0004ss00990.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015043 | 0.0 | AP015043.1 Vigna angularis var. angularis DNA, chromosome 10, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022643111.1 | 0.0 | two-component response regulator ORR21 isoform X2 | ||||
| Swissprot | Q8H7S7 | 1e-123 | ORR21_ORYSJ; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A3Q0FHC8 | 0.0 | A0A3Q0FHC8_VIGRR; Two-component response regulator | ||||
| STRING | XP_007153220.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11808 | 24 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-95 | response regulator 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



