 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang0045ss01380.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1133aa MW: 128111 Da PI: 5.6638 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179.2 | 5.1e-56 | 48 | 164 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenpt 93
++ ++rwl++ ei+aiL n++k++++ e++t p+sgsl+L++rk++ryfrkDG++w+kkkdgktvrE+he+LK g+v+vl+cyYah+ee+++
Vang0045ss01380.1 48 VEAQHRWLRPAEICAILSNYKKFRIAPEPATMPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVREAHERLKAGSVDVLHCYYAHGEEDES 139
5679**************************************************************************************** PP
CG-1 94 fqrrcywlLeeelekivlvhylevk 118
fqrr+ywlLeee+++ivlvhy++vk
Vang0045ss01380.1 140 FQRRTYWLLEEEHSNIVLVHYRQVK 164
**********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 77.835 | 43 | 169 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.2E-75 | 46 | 164 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.2E-48 | 49 | 162 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.9E-15 | 549 | 634 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 4.8E-5 | 549 | 625 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 2.18E-19 | 729 | 845 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.36E-16 | 731 | 843 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 4.2E-19 | 731 | 847 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.863 | 734 | 855 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 3.6E-9 | 756 | 851 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.965 | 784 | 816 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.002 | 784 | 813 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 390 | 823 | 852 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.38E-7 | 951 | 1006 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.32 | 956 | 978 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 958 | 986 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0046 | 958 | 977 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.038 | 979 | 1001 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.62 | 980 | 1004 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.001 | 982 | 999 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1133 aa Download sequence Send to blast |
MAEARQYFPP SQLGQLFFSL IPLLLLEIPI IQTLKKETLF RNIKQIIVEA QHRWLRPAEI 60 CAILSNYKKF RIAPEPATMP PSGSLFLFDR KVLRYFRKDG HNWRKKKDGK TVREAHERLK 120 AGSVDVLHCY YAHGEEDESF QRRTYWLLEE EHSNIVLVHY RQVKGTKANY TCAKENEESL 180 PCAEQTDKIM LKAEMDTSFS SALHPNSYQV PSQTMDSSMN SAQVSEYEEA ESAFNSHASS 240 EFYSFLELQR PVEKIIDQPA DSYSPHPLIN EQKKLPVIAD MNYISLTQDR KFIDIHNGGL 300 IYESPKPLGF SSWEDILENN GESQHMPFQP LFPEMQPDEM AVNSSFCQGY DIMVPHLTTS 360 IAKLHDNGSL IQAEGSWQGY SVDSLRVSSW PIDNVHSSST CEVSCSNCEH EVNEVDIQKS 420 LQQSLLHQHK QNKVLMQNDP QEIMLDYLKS DFEASRTLDG IEDPRFTFKR TLLDGSPAEE 480 GLKKLDSFNQ WMSKELGDVE ESNKPSTSGA YWDTVESESE VGSTTIPSQG HLDTYVLDPS 540 VSNEQLFSII DYSPSWAFEG SEVKIIISGR FLRSQQEAEQ CKWSCMFGEV EVPAVILAKD 600 VLCCHTPLHK AGRVPFYVTC SNRLACSEVR EFDFQVNCTQ EVNTAGEDRA IVFHTLSRRF 660 GELLSLGHAV PQNSDSISEN EKSELRSKIS SLLRGEDDVW DKLLELTQEK DISPEDLQEQ 720 LLQNLLKDKL HAWLLQKITD DGKGPNVLDE GGQGVLHFAA ALGYDWALEP TLVAGVNVNF 780 RDINGWTALH WAAFYGRERT VAFLISLGAA PGLLTDPCPE HPSGRTPADL ASTNGHKGIA 840 GYLAESSLSA QLTTLDLNKD VGENSGNKVV QRIQDIAPVN DLDVLSYEQS LKDSLAAVCN 900 ATQAAARIHQ VFRMQSFQRK QLKEFGDDKF GISDERALSL VKKNGKSHKS GSRDEPVHAA 960 AIRIQNKFRG WKGRKDFLMI RQRIVKIQAH VRGHQVRKNC GKIIWSVGIL EKVILRWRRK 1020 GSGLRGFKSE ANSEVTMIED VTSSEDHYDV LKEGRKQTEQ RLQKALARVK SMIQYPEARD 1080 QYHRVLNVVT QIQDNQVKHD SSCNNSEEIR DFNNLTDLEA LLDEDIFMPT AT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ydw_A | 1e-11 | 683 | 843 | 12 | 165 | DARPIN 44C12V5 |
| 4ydw_B | 1e-11 | 683 | 843 | 12 | 165 | DARPIN 44C12V5 |
| 4ydy_A | 1e-11 | 683 | 843 | 12 | 165 | DARPIN 44C12V5 |
| 4ydy_B | 1e-11 | 683 | 843 | 12 | 165 | DARPIN 44C12V5 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
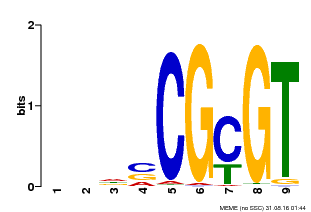
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang0045ss01380.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 0.0 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017442435.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A0S3QYK9 | 0.0 | A0A0S3QYK9_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007159660.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8284 | 27 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||



