 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang0155s00070.1 | ||||||||
| Common Name | LR48_Vigan08g092500 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 175aa MW: 20492.3 Da PI: 9.1494 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 116.6 | 1.5e-36 | 4 | 102 | 1 | 99 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkae 93
aCaaCk++r+kC+++C+lapyfp++++++f +vhk+FG+sn++kl+k+++e++r+++++sl++eA++r+rdP++G +g ++k+ ++++++ e
Vang0155s00070.1 4 ACAACKHQRKKCSEKCILAPYFPSNRSREFHAVHKVFGVSNITKLVKNAREDDRRRVVDSLTWEACCRQRDPIHGPYGEYTKVFNEYKKVLDE 96
7*****************************************************************************************999 PP
DUF260 94 lallke 99
l++ ++
Vang0155s00070.1 97 LKRFRS 102
998776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 23.82 | 3 | 104 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 2.2E-34 | 4 | 100 | IPR004883 | Lateral organ boundaries, LOB |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 175 aa Download sequence Send to blast |
MHPACAACKH QRKKCSEKCI LAPYFPSNRS REFHAVHKVF GVSNITKLVK NAREDDRRRV 60 VDSLTWEACC RQRDPIHGPY GEYTKVFNEY KKVLDELKRF RSQHQMLQFP SLGFKSVQDK 120 AEEDYLHGKK NAIIDSDIYN SYSSNYLQEF QNLRPEVLIP FHHHSQPYYI TGIL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 3e-19 | 5 | 97 | 12 | 104 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 3e-19 | 5 | 97 | 12 | 104 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
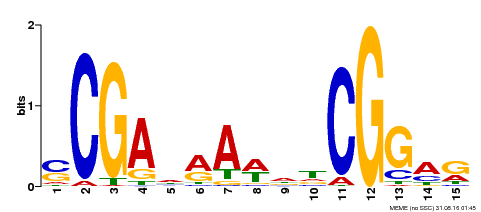
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00128 | DAP | Transfer from AT1G06280 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang0155s00070.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015039 | 0.0 | AP015039.1 Vigna angularis var. angularis DNA, chromosome 6, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014524391.2 | 1e-123 | LOB domain-containing protein 2 | ||||
| Swissprot | Q9LNB9 | 6e-38 | LBD2_ARATH; LOB domain-containing protein 2 | ||||
| TrEMBL | A0A0L9V574 | 1e-129 | A0A0L9V574_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007132691.1 | 1e-111 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12722 | 25 | 34 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06280.1 | 3e-40 | LOB domain-containing protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



