 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang01g02950.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 416aa MW: 47438.3 Da PI: 9.2216 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.9 | 7.5e-05 | 78 | 97 | 2 | 21 |
EETTTTEEESSHHHHHHHHH CS
zf-C2H2 2 kCpdCgksFsrksnLkrHir 21
+C+ Cg +F+ + +L +H++
Vang01g02950.1 78 TCEECGVTFKKHAYLLQHMQ 97
6******************7 PP
| |||||||
| 2 | zf-C2H2 | 18.2 | 7.1e-06 | 160 | 184 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ dC+ s++rk++L+rH+ +H
Vang01g02950.1 160 FVCTvdDCHASYRRKDHLTRHLLLH 184
89********************998 PP
| |||||||
| 3 | zf-C2H2 | 13.4 | 0.00023 | 189 | 214 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp +C+ Fs + n++rH H
Vang01g02950.1 189 FKCPvvNCNIEFSLQGNMQRHVEEiH 214
89*******************98777 PP
| |||||||
| 4 | zf-C2H2 | 17.7 | 1e-05 | 229 | 253 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp Cgk F+ s+Lk+H +H
Vang01g02950.1 229 HVCPeiGCGKGFRFASQLKKHEDSH 253
79*******************9888 PP
| |||||||
| 5 | zf-C2H2 | 14 | 0.00015 | 320 | 345 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C+ C+ +Fs ksnL +H + H
Vang01g02950.1 320 FQCEfkGCSCTFSKKSNLDKHKKAvH 345
89*******************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 28 | 33 | 56 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 26 | 77 | 97 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-9 | 157 | 186 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.49E-6 | 158 | 186 | No hit | No description |
| PROSITE profile | PS50157 | 10.221 | 160 | 189 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 160 | 184 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 162 | 184 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.28E-5 | 187 | 215 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.4E-4 | 187 | 215 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.159 | 189 | 219 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0054 | 189 | 214 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 191 | 214 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-6 | 223 | 253 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.408 | 229 | 258 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.016 | 229 | 253 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 231 | 253 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.92 | 261 | 286 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-4 | 262 | 286 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 263 | 286 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 35 | 289 | 310 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.4E-7 | 301 | 342 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.42E-9 | 304 | 359 | No hit | No description |
| PROSITE profile | PS50157 | 13.609 | 320 | 350 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0098 | 320 | 345 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 322 | 345 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-5 | 344 | 374 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.7 | 351 | 377 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 353 | 377 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 416 aa Download sequence Send to blast |
LHSSICSLYL ITKTMVEAVT IEERPKSRDI RRYFCQYCGI CRSKKTLITS HINSQHKEED 60 EKARKETDPE TESSKSNTCE ECGVTFKKHA YLLQHMQNSK IIGKLWINSV KHWIPDNELQ 120 NARGNEFRLD GSVSGRETSC EAPSRMPQSR SLNLRLARPF VCTVDDCHAS YRRKDHLTRH 180 LLLHEGKTFK CPVVNCNIEF SLQGNMQRHV EEIHDGSSTA AADKSHKQHV CPEIGCGKGF 240 RFASQLKKHE DSHVKLESIE VVCLEPGCMK HFANVQGLRA HVNTSHQYMT CDICGSKQLK 300 KNIKRHLRSH EADSSSSKTF QCEFKGCSCT FSKKSNLDKH KKAVHLKEKP FRCGFPDCGM 360 RFAYKHVRDN HEKTAKHVFT LVSSFSFWVI LKKLMQNSGQ GRGEGGRENV LLLKC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2i13_A | 2e-13 | 158 | 374 | 20 | 183 | Aart |
| 2i13_B | 2e-13 | 158 | 374 | 20 | 183 | Aart |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
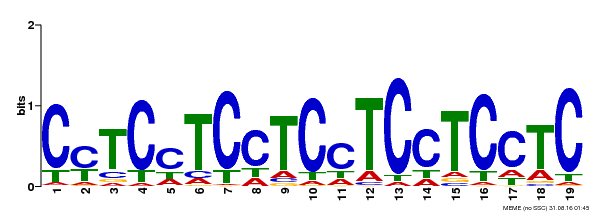
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang01g02950.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015036 | 1e-160 | AP015036.1 Vigna angularis var. angularis DNA, chromosome 3, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017429048.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| TrEMBL | A0A0L9UUX0 | 0.0 | A0A0L9UUX0_PHAAN; Uncharacterized protein | ||||
| TrEMBL | A0A0S3RJL2 | 0.0 | A0A0S3RJL2_PHAAN; Uncharacterized protein | ||||
| STRING | XP_004498336.1 | 1e-149 | (Cicer arietinum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-100 | transcription factor IIIA | ||||



