 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang02g02820.1 | ||||||||
| Common Name | LR48_Vigan10g045200 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 504aa MW: 54780 Da PI: 5.4071 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.2 | 7e-17 | 83 | 127 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G g W+ I +++g ++t+ q++s+ qk+
Vang02g02820.1 83 REKWTEEEHQKFLEALKLYGRG-WRQIEEHIG-TKTAVQIRSHAQKF 127
789*******************.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.26E-16 | 77 | 131 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.201 | 78 | 132 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.5E-10 | 80 | 134 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.1E-16 | 81 | 130 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 4.5E-13 | 82 | 130 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-14 | 83 | 126 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.99E-11 | 85 | 128 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 504 aa Download sequence Send to blast |
LSLLAAGVSL CGGLGCFPRQ ADSHFAPMEM QDQIENTRST ISGSASNCHS NAGKQSENVA 60 HIPSVRNGHA PKVRKPYTIT KQREKWTEEE HQKFLEALKL YGRGWRQIEE HIGTKTAVQI 120 RSHAQKFFSK VVRESEGSAE SSIQPINIPP PRPKRKPLHP YPRKSVDAFR GHAIPNETET 180 SQSTNLLVAE KDTPSPTSVL STVGSEAFGS AFSEQNNRCL SPNSCTTDIH SVSLSPVEKE 240 NDCMTSKASE EEEKGSPASV PLSIVSDPNM CMKPEFSSKE TEYGIDDSAN MPQTTSIKLF 300 GRTVSMVDNQ KPQNIDDNGK AIITVKSDVV DDAGNEKLGQ STEQVDTQLS LGLVGGNCSI 360 TPDSDGADRS EEPPKENVCL SECVPDASFP QWSLYQGLPA FCVSPCNQIL NHMPLRPSLK 420 VRAREEESCC TGSNTESVCD MENQGKNSDA VDSKCEKYEG EGAAPQKPAR GFVPYKRCLA 480 ERDGTSLIAA LEERDGQRAR VCS* |
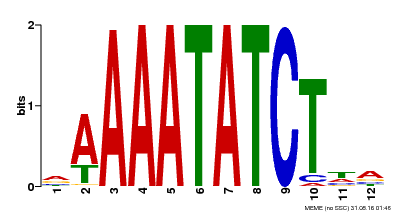
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang02g02820.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015044 | 0.0 | AP015044.1 Vigna angularis var. angularis DNA, chromosome 11, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017439841.1 | 0.0 | PREDICTED: protein REVEILLE 7-like | ||||
| Refseq | XP_017439842.1 | 0.0 | PREDICTED: protein REVEILLE 7-like | ||||
| TrEMBL | A0A0L9VHM4 | 0.0 | A0A0L9VHM4_PHAAN; Uncharacterized protein | ||||
| TrEMBL | A0A0S3TC58 | 0.0 | A0A0S3TC58_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007152597.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7313 | 32 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.2 | 1e-52 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



