 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang03g00950.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 523aa MW: 58060.3 Da PI: 5.1656 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 58 | 1.7e-18 | 97 | 181 | 4 | 83 |
E-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 4 vltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
+lt sd++++g +++ +++a+++ ++++ + +l+ +d++g++W +++i+r++++r++lt+GW+ Fv++++L +gD ++F
Vang03g00950.2 97 TLTASDTSTHGGFSVLRRHADDClppldmtQQPPWQ--ELVATDLHGNEWHFRHIFRGQPRRHLLTTGWSVFVSSKKLVAGDAFIFL 181
6899***********************975444444..8***********************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01019 | 1.3E-9 | 95 | 198 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 1.06E-32 | 97 | 189 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 2.9E-30 | 97 | 183 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 1.8E-16 | 97 | 181 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.58E-15 | 97 | 180 | No hit | No description |
| PROSITE profile | PS50863 | 9.234 | 133 | 197 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02309 | 4.8E-6 | 396 | 434 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 26.402 | 398 | 491 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 9.16E-11 | 399 | 475 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 523 aa Download sequence Send to blast |
MASAAPGSTN DALYKELWHA CAGPLVTLPR EGVRVYYFPQ GHMEQLEASM NQGLEQQMPS 60 FNLPSKILCK VVNVHLRAEP ETDEVYAQIT LLPEADTLTA SDTSTHGGFS VLRRHADDCL 120 PPLDMTQQPP WQELVATDLH GNEWHFRHIF RGQPRRHLLT TGWSVFVSSK KLVAGDAFIF 180 LRQVVQWDEP SSILRPDRVS PWELEPLVST PPTNSQPSQR NKRSRPPILP STLADSSLQG 240 VRKSPVDSAP FSYRDHQHGG DIYPPAKFNS TATGFLGFGG NCSASNKSIY WSSRTENSTE 300 SFLPVALKES GEKRNVTANG CRLFGIQLLD NSNGEESLPM VTLSGRLVDD VPLPSLDAES 360 DQHSEPSNIN RSDIPSVSCD VEKSCLRSPH ESQSRQIRSC TKVHMQGMAV GRAVDLTRFD 420 GYEDLLRKLE EMFDIKDELC GSPKKWQVVY TDNEDDMMMA GDDPWLEFCS IVRKIFIYTA 480 EEVKKLSPKI GLPISEEVKP SKMDSEAVVN PEDQSSIVGS GC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldy_A | 1e-110 | 2 | 182 | 7 | 212 | Auxin response factor 1 |
| 4ldy_B | 1e-110 | 2 | 182 | 7 | 212 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Acts as repressor of IAA2, IAA3 and IAA7. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16176952}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | Transfer from AT1G59750 | Download |
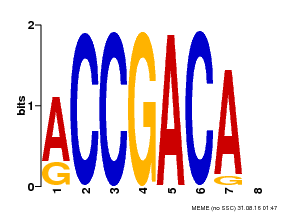
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang03g00950.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015038 | 0.0 | AP015038.1 Vigna angularis var. angularis DNA, chromosome 5, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021888215.1 | 0.0 | LOW QUALITY PROTEIN: auxin response factor 1-like | ||||
| Swissprot | Q8L7G0 | 1e-107 | ARFA_ARATH; Auxin response factor 1 | ||||
| TrEMBL | A0A444ZC76 | 0.0 | A0A444ZC76_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007148592.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59750.4 | 1e-111 | auxin response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



