 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang04g00270.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 527aa MW: 57590.2 Da PI: 7.9453 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 104.9 | 4.4e-33 | 224 | 281 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgse+prsYY+Ct+ +C vkkkver+ d++++ei+Y+g+Hnhek
Vang04g00270.1 224 DDGYNWRKYGQKQVKGSEYPRSYYKCTHLNCLVKKKVERAP-DGHITEIIYKGQHNHEK 281
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 107.2 | 8.1e-34 | 412 | 470 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+CtsagC+v+k+ver+++dpk+v++tYeg+Hnh+
Vang04g00270.1 412 LDDGYRWRKYGQKVVKGNPHPRSYYKCTSAGCNVRKHVERASTDPKAVITTYEGKHNHD 470
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 8.0E-29 | 211 | 283 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.789 | 218 | 282 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.41E-26 | 222 | 282 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-36 | 223 | 281 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.8E-25 | 224 | 280 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.5E-37 | 397 | 472 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.31E-29 | 404 | 472 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 39.192 | 407 | 472 | IPR003657 | WRKY domain |
| SMART | SM00774 | 9.6E-40 | 412 | 471 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.9E-26 | 413 | 470 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:1900150 | Biological Process | regulation of defense response to fungus | ||||
| GO:1900425 | Biological Process | negative regulation of defense response to bacterium | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 527 aa Download sequence Send to blast |
MSTPNADSGM APHPRPTITL PPRPSAEAFF SAAGGASPGP MTLVSSFFGS DAAADCRSFS 60 QLLAGAMASP MAFSATVADN SDKDDDGPHK GFKQSRPMNL VIARSPVFTV PPGLSPSGFL 120 NSPGFFSPQS PFGMSHQQAL AQVTAQAVLA QSHMHMQADY QMHPVTAPTE PPVQQPSFAL 180 NEASEQKIVP SVSEAKNAQL ETSDISQADK KYQLASQAID KPADDGYNWR KYGQKQVKGS 240 EYPRSYYKCT HLNCLVKKKV ERAPDGHITE IIYKGQHNHE KPQANRRVKD NSDSNGSAIV 300 QPKSESNSQG WVGQQVNKFS ENIPDCSVPE SDQISNQGAP RQLLPGSSGS EEVGDVDNRE 360 EADDIEPNPK RRQAKFLLFR NTDVAVSEVP LSQKTVTEPK IIVQTRSEVD LLDDGYRWRK 420 YGQKVVKGNP HPRSYYKCTS AGCNVRKHVE RASTDPKAVI TTYEGKHNHD VPAARNSSHN 480 TANTNSVPLK PHNVVPEKHP LLKDMDFGSN DQRPVHLRLK EEQIIV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-40 | 224 | 473 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-40 | 224 | 473 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Has a positive role in resistance to necrotrophic pathogens (e.g. Botrytis cinerea), but a negative effect on plant resistance to biotrophic pathogens (e.g. Pseudomonas syringae). {ECO:0000269|PubMed:18570649, ECO:0000269|PubMed:22219184}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
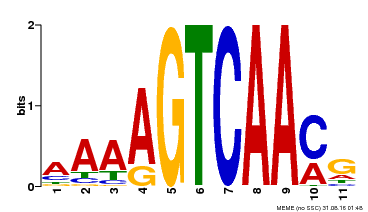
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang04g00270.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By biotic and abiotic stresses such as pathogen infection (e.g. Botrytis cinerea and Pseudomonas syringae), salicylic acid (SA), jasmonic acid (JA), ethylene (ACC), liquid infiltration or spraying, and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:18570649}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 0.0 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017418807.1 | 0.0 | PREDICTED: probable WRKY transcription factor 4 | ||||
| Swissprot | Q9XI90 | 1e-143 | WRKY4_ARATH; Probable WRKY transcription factor 4 | ||||
| TrEMBL | A0A0S3RRX9 | 0.0 | A0A0S3RRX9_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007139560.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1545 | 34 | 100 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13960.1 | 1e-125 | WRKY DNA-binding protein 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



