 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang04g00390.1 | ||||||||
| Common Name | LR48_Vigan03g021600 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 244aa MW: 27860.6 Da PI: 8.7275 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.2 | 2.9e-16 | 24 | 71 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++ Ed ll ++v+++G g W++++++ g++R++k+c++rw++yl
Vang04g00390.1 24 KGAWSQTEDGLLRECVQLYGEGKWHLVPQRAGLNRCRKSCRLRWLNYL 71
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 46.3 | 9.9e-15 | 77 | 121 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg ++++E +l ++++k+lG++ W++Ia +++ gRt +++k++w++
Vang04g00390.1 77 RGDFSEDEVDLMIRLHKLLGNR-WSLIAGRLP-GRTSNDVKNYWNTN 121
899*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.664 | 19 | 71 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.37E-27 | 21 | 118 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-21 | 21 | 74 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.5E-13 | 23 | 73 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.2E-15 | 24 | 71 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.65E-8 | 26 | 71 | No hit | No description |
| PROSITE profile | PS51294 | 24.073 | 72 | 126 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-22 | 75 | 126 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.9E-14 | 76 | 124 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.3E-13 | 77 | 120 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.88E-9 | 80 | 122 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 244 aa Download sequence Send to blast |
MYQVMTSVKA ACVMMEGRVS GVRKGAWSQT EDGLLRECVQ LYGEGKWHLV PQRAGLNRCR 60 KSCRLRWLNY LKPNIKRGDF SEDEVDLMIR LHKLLGNRWS LIAGRLPGRT SNDVKNYWNT 120 NMRRKVQSHN RDEENNNNNV KESERSWKPH KVIKPVPRAL AKASAMVHGK LMSSSKVGVS 180 EAVSAGSENW WETLLDDKEE NINNSTCFPV GKDGSFELWD EELVSIPSEF FAEAETWSDF 240 LLN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 7e-25 | 19 | 128 | 2 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | Transfer from AT1G66370 | Download |
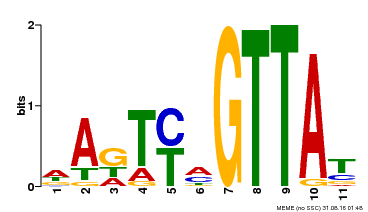
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang04g00390.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 0.0 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017416381.1 | 1e-168 | PREDICTED: transcription factor MYB90-like | ||||
| Swissprot | Q9FNV9 | 3e-56 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | A0A0L9U222 | 1e-177 | A0A0L9U222_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007139536.1 | 1e-136 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66370.1 | 3e-57 | myb domain protein 113 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



