 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang04g16730.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 358aa MW: 40668.5 Da PI: 5.4971 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.3 | 1.3e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT+eEde+l ++++ G g+W+t ++ g+ R++k+c++rw +yl
Vang04g16730.1 14 KGRWTAEEDEILTKYIQANGEGSWRTLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.3 | 5.4e-16 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg + eE+ +v++++ lG++ W++Ia++++ gRt++++k++w+++l
Vang04g16730.1 67 RGDISLEEENTIVKLHASLGNR-WSLIASHLP-GRTDNEIKNYWNSHL 112
567789****************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-21 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.863 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.06E-27 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.7E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.5E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.84E-8 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 24.812 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.0E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.19E-9 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0051555 | Biological Process | flavonol biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 358 aa Download sequence Send to blast |
MGRAPCCEKV GMKKGRWTAE EDEILTKYIQ ANGEGSWRTL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGDI SLEEENTIVK LHASLGNRWS LIASHLPGRT DNEIKNYWNS HLSRKIYTFR 120 GTTNSKPDII TLTLPPKRKR GRTSRWAMKK NKTYCAPKQK QPLLPAVPLP PTPPLETEKH 180 SSTHENTTTQ EEENLVTNAI WVEGEKEIMN IWGPYDEEVI NGGEGDQVLS FESFNDIMNS 240 YCSQASGCWT ISDEGYSESS EVVGNKTCDH TVLSSGREKQ CFCSTKFSSD SDLCQSAAQL 300 DHKDESLLDH KDESVSWEQH QSLLSWLCED EDWEKDLQSV GEIDPQKQKD MVDWFLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 1e-22 | 14 | 116 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 4e-22 | 14 | 116 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 4e-22 | 14 | 116 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 3e-22 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 1e-22 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-22 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
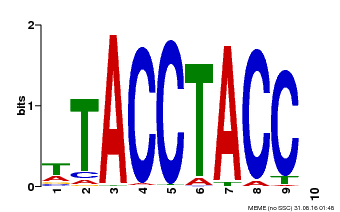
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang04g16730.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 0.0 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017418017.1 | 0.0 | PREDICTED: transcription factor MYB12-like isoform X1 | ||||
| Refseq | XP_017418018.1 | 0.0 | PREDICTED: transcription factor MYB12-like isoform X2 | ||||
| TrEMBL | A0A0S3S0A3 | 0.0 | A0A0S3S0A3_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007163163.1 | 1e-154 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49330.1 | 2e-75 | myb domain protein 111 | ||||



