 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang05g01960.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 253aa MW: 27698.9 Da PI: 10.104 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 212.1 | 1.4e-65 | 17 | 154 | 2 | 143 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqs 96
+++rkp+w+ErEnn+rRERrRRaiaakiy+GLRaqGny+lpk++DnneVlkALc+eAGw+ve+DGttyrkg++ + a+ +g+s++++p ss++
Vang05g01960.1 17 ANRRKPSWRERENNRRRERRRRAIAAKIYTGLRAQGNYNLPKHCDNNEVLKALCAEAGWTVEEDGTTYRKGCRAP-LADGVGTSTRNTPFSSQNP 110
679**********************************************************************99.9****************** PP
DUF822 97 slkssalaspvesysaspksssfpspssldsislasaasllpvlsvl 143
s+ ss+++sp++sy+ sp+sssfpsps+ld ++++ +l+p++++
Vang05g01960.1 111 SPLSSSFPSPIPSYQVSPSSSSFPSPSRLDDNNAS---NLIPYIRHS 154
********************************886...455554443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 7.9E-62 | 18 | 146 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 253 aa Download sequence Send to blast |
LAELAMTSDG ATSAAVANRR KPSWRERENN RRRERRRRAI AAKIYTGLRA QGNYNLPKHC 60 DNNEVLKALC AEAGWTVEED GTTYRKGCRA PLADGVGTST RNTPFSSQNP SPLSSSFPSP 120 IPSYQVSPSS SSFPSPSRLD DNNASNLIPY IRHSFSASLP PLRISNSAPW LNFQAFGPSV 180 SAVPISPTLN FIKPVPSQQH KHNLNHPDNR IQEMRNSEVQ FGVQVKPWVG ERIHEGGLDD 240 LELTLGSGKT LA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 2e-31 | 19 | 101 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 2e-31 | 19 | 101 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 2e-31 | 19 | 101 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 2e-31 | 19 | 101 | 371 | 453 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 18 | 37 | RRKPSWRERENNRRRERRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive regulator of brassinosteroid (BR) signaling. Transcription factor that activates target gene expression by binding specifically to the DNA sequence 5'-CANNTG-3'(E box) through its N-terminal domain. Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BIM1, BIM2 or BIM3. The C-terminal domain is probably involved in transcriptional activation (PubMed:12007405, PubMed:15680330, PubMed:18467490, PubMed:19170933). Recruits the transcription elongation factor IWS1 to control BR-regulated gene expression (PubMed:20139304). Forms a trimeric complex with IWS1 and ASHH2/SDG8 to regulate BR-regulated gene expression (PubMed:24838002). Promotes quiescent center (QC) self-renewal by cell divisions in the primary root. Binds to the E-boxes of the BRAVO promoter to repress its expression (PubMed:24981610). {ECO:0000269|PubMed:12007405, ECO:0000269|PubMed:15680330, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:19170933, ECO:0000269|PubMed:20139304, ECO:0000269|PubMed:24838002, ECO:0000269|PubMed:24981610}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
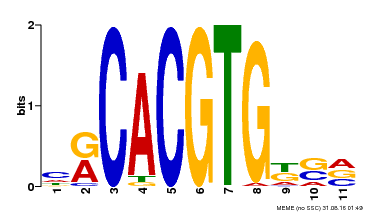
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang05g01960.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015035 | 1e-142 | AP015035.1 Vigna angularis var. angularis DNA, chromosome 2, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028803037.1 | 7e-95 | protein BRASSINAZOLE-RESISTANT 1-like | ||||
| Swissprot | Q9LN63 | 2e-53 | BZR2_ARATH; Protein BRASSINAZOLE-RESISTANT 2 | ||||
| TrEMBL | A0A151TBZ7 | 1e-100 | A0A151TBZ7_CAJCA; Protein BRASSINAZOLE-RESISTANT 1 | ||||
| STRING | XP_008444689.1 | 2e-77 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1275 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.6 | 3e-41 | BES1 family protein | ||||



