 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vang0974s00030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 324aa MW: 37003.9 Da PI: 8.0704 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 21.6 | 5.1e-07 | 64 | 110 | 2 | 46 |
SSS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
g WT+eE +l+ +a + + ++ +W +a++++ g+t +++ ++
Vang0974s00030.1 64 GGWTAEENKLFENALAYYDKDtpdRWMMVAAMIP-GKTVGDVIQQYRE 110
56********************************.*******999986 PP
| |||||||
| 2 | Myb_DNA-binding | 46 | 1.2e-14 | 172 | 216 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + k++G+g+W+ I+r + ++Rt+ q+ s+ qky
Vang0974s00030.1 172 PWTEEEHRQFLLGLKKYGKGDWRNISRNFVTTRTPTQVASHAQKY 216
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 8.438 | 61 | 116 | IPR017884 | SANT domain |
| SMART | SM00717 | 3.4E-6 | 62 | 114 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.64E-11 | 64 | 119 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.50E-5 | 66 | 111 | No hit | No description |
| PROSITE profile | PS51294 | 20.244 | 165 | 221 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.28E-17 | 166 | 220 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.9E-18 | 168 | 220 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.5E-13 | 169 | 219 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-12 | 169 | 216 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.4E-12 | 172 | 216 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.40E-10 | 172 | 217 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
MLCPSLILFS FRTKSLWIWI PLSNISSYPV HSYLGEIMNR AIEVLSPASY LQNSNWLFQE 60 SQGGGWTAEE NKLFENALAY YDKDTPDRWM MVAAMIPGKT VGDVIQQYRE LEEDVSVIEA 120 GLIPVPGYTT SSFTLEWVNS QSYDEFKHFC TVGGKRGAST RPSEQERKKG VPWTEEEHRQ 180 FLLGLKKYGK GDWRNISRNF VTTRTPTQVA SHAQKYFIRQ LTGGKDKRRS SIHDITMVNL 240 QETKSPSSDT TRPSSPLQNQ KLSSMVKQEY DWKSPCDDMP FVFNSTNGNM FMAAPLHGIS 300 SYESRPQEHN ILRQSPWMPV FTL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 3e-16 | 63 | 131 | 8 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
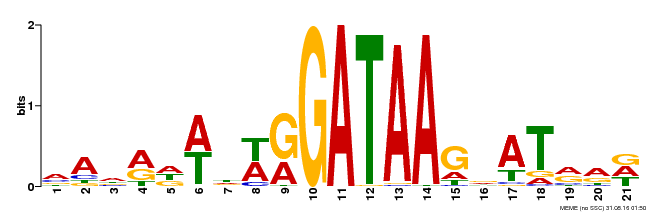
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vang0974s00030.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 0.0 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017419677.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 1e-105 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A0S3RU74 | 0.0 | A0A0S3RU74_PHAAN; Uncharacterized protein | ||||
| STRING | XP_007140381.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF466 | 34 | 162 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 1e-115 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



