 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0001s0782.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1394aa MW: 139822 Da PI: 4.4455 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.6 | 1.4e-13 | 16 | 71 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT........TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg........kgRtlkqcksrwqkyl 48
+g WT+eEd l+++v ++G g+W+ Iar ++ +gR +kqc++rw+ +l
Vocar.0001s0782.1.p 16 KGGWTAEEDAHLARLVLEYGEGNWSPIARALNvlmknpesTGRIGKQCRERWNHHL 71
688*************************************************9886 PP
| |||||||
| 2 | Myb_DNA-binding | 54.5 | 2.7e-17 | 77 | 119 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+g+W +eE+ ll+da+++lG++ W+ Iar+++ gR+++ +k++w+
Vocar.0001s0782.1.p 77 KGPWGPEEEILLADAHRRLGNK-WSDIARCIP-GRSENAVKNHWN 119
79********************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.365 | 11 | 71 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.47E-26 | 14 | 118 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-11 | 15 | 73 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.8E-12 | 16 | 70 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-19 | 18 | 78 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.83E-9 | 19 | 71 | No hit | No description |
| PROSITE profile | PS51294 | 22.187 | 72 | 126 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.0E-14 | 76 | 124 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-15 | 77 | 119 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.93E-10 | 79 | 122 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-19 | 79 | 126 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1394 aa Download sequence Send to blast |
MSKRCAGPRH PIAKQKGGWT AEEDAHLARL VLEYGEGNWS PIARALNVLM KNPESTGRIG 60 KQCRERWNHH LSPGLRKGPW GPEEEILLAD AHRRLGNKWS DIARCIPGRS ENAVKNHWNA 120 TLRKRLLDNE VGGPLKQYLD TLNLAVPPRK RSGGSGGKGG QGGGRRTKAR RASREREDSG 180 SDDASEGLGG LEDCDSDAPE PTRRRRPAAR GGRGSMRALD ADGGRGGAGG SSSGAVGGDL 240 NLGASAAAGR SWPPSPATRR GGAGSSGGAS GSGGGGRGVN SRGGVAPLGG ARRTGGSSGA 300 GGNSTPRGNG GISDAGGGGG IIGGRDSDGV PIGASAAVQA PVQARTAPKR LSCNGLGSGA 360 TGGGTIIKLG PYSAAAAARA AEMAKARAAA DAAAAAAIGN YRRRRSVRAA AQVGNERLRS 420 LAEAEQREAE EAAEDADAAA ADAADAAAAS AEPPVKAEVK LEEDYLGDID FQLDDETLDD 480 LFAYGGASCS DRDADRDAAE SVGDGVDDDY DDDDGDGDND DDDGDNDDDD DDNEEDEEEE 540 DDFYLESDGE CDGGAGEAEM SDMQLSVSGL VPDGGANPCP GAATATATAA AASIAASAAV 600 AAAAAAAAMS AVAAKRRSSS GFPSTTSAAA AAAAAAAALL PDSFLAAKPP AAAAAGGGRP 660 DGGGGDDVDG ELYGRAAPIR SLRRGAERAF DNSPVQVLAV QKARRTSYGS GAAGTARLSA 720 FDAASRRVSS ADGFGAAAIG DESADALTAA INAAAAAAAA VSADETPFRY VRKEEGANAP 780 GDEGLMAASN DAFVLAGAGA LCHVASDSAT VNMRPSADGT GGTLPLSAWA SPPPLNPRNS 840 SGDLGAAAAS AAMPAALLLL PPPPPPANIH IHSPGLHDLL DFDGMVSPLA AAVGGALEEQ 900 VDGAVAAAAT MATGGGGGGS RVGSGGGGMD SFAGTLNNVA DISSGITSVG SPVSDLGDIF 960 DSYIAFGGIE DAKLCTGATC EEAAAAAVAV AAAAAADGGQ ADNEILGAGD IMYGGVSLKR 1020 ESSFTGCGLR PLLPPSMESS SAGNQFQISN FLATFQGPAA PALPPPPAAP APPLQLPWTK 1080 MGEPAAAAAP PPPPPPPLLS AVAITPASPP VPWPAVPTQQ GGIPVAPMDM PYVAGGGSSG 1140 AMSPLPGLTS TGLLQPTSPF AWLGQQPAAS VLQPPPPPVQ QMQMQAMYGG GAPAVQLPPP 1200 SGLITQMQQQ QAVQAQEQQQ PQQSAERVSS YPVGIGAASY PGGSNMPFGG MYGALPYPPA 1260 VGHPQAVAVP VPVPVPVPQM DQQYMQQYMQ QYMQQYMQQM HIQMHQALLQ QLSLPPDPQP 1320 ASQSTGIKVP EVNLLEKSNF RLNLILDMEA MRTIATKVEG SSLHPVGFCA QPPADNNNNN 1380 DNDNDNAGNA ATR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 5e-36 | 16 | 126 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 3e-35 | 16 | 126 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 3e-35 | 16 | 126 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 5e-36 | 16 | 126 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 5e-36 | 16 | 126 | 4 | 105 | C-Myb DNA-Binding Domain |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 656 | 664 | GGRPDGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
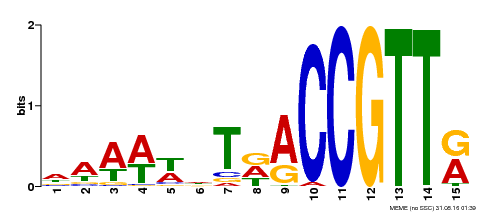
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 3e-34 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0001s0782.1.p |



