 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0003s0248.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 823aa MW: 81750.8 Da PI: 4.6772 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54.1 | 2.1e-17 | 56 | 85 | 1 | 30 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyy 30
C+ Cg+t+Tp+WR+gp g+ktLCnaCG++y
Vocar.0003s0248.2.p 56 CVECGATQTPQWREGPAGPKTLCNACGVRY 85
****************************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50114 | 12.479 | 50 | 85 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 1.8E-11 | 50 | 106 | IPR000679 | Zinc finger, GATA-type |
| Gene3D | G3DSA:3.30.50.10 | 3.8E-14 | 53 | 88 | IPR013088 | Zinc finger, NHR/GATA-type |
| SuperFamily | SSF57716 | 5.23E-12 | 53 | 90 | No hit | No description |
| CDD | cd00202 | 2.09E-12 | 55 | 105 | No hit | No description |
| Pfam | PF00320 | 8.2E-15 | 56 | 85 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 56 | 81 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 823 aa Download sequence Send to blast |
MEVEDGRRHG KRSQAGISGH VEGVADSEEV KRQRVAEEQP AAPVRRTGKL VSGRTCVECG 60 ATQTPQWREG PAGPKTLCNA CGVRYVRAQQ RANKRAVPAG AARSGYGAGG RQSRTSKAAK 120 AEVASARAAR TAAVVPAAAA GMAPGEEASQ RPQRQAALMA ASRTAQYART GFFPLDTIDL 180 QQQQQQHTPD NAAINGQIAM HHQHHQHEAD MGDVLTGAFG GVSSAGAAAA GAAAAGPGSP 240 NCSSYDSCGS LQEGVTVEVV VGAADGEAEC CGGSVAAVAP AAAAAATAAT VPTYPLLTSS 300 SVGAAAGSLM PTPAQLIGGS GIMLGGVEES VMAVAPVMAL PLGEGQLQSA VPGALQTASA 360 VAAAVAAAGV LGTASSEFVL ECDTTGSGMF FASGLDCQFS GDGVCYPADS HHHHHHHHDD 420 HDQNGVCHHS KTGCVAPAPS GGQCDTAAVY QGTGAVSCVA QVKSVPVSPC VDVDGAGFGC 480 GGFGSVSRML DGVLASARGY GSQLGCGLVG GLDDEYTPLV EVEVDFGLGP ASAMGDGSLF 540 DGFDSGTCGA DTVEDDDPQH PQSQPLLAQG PALSASQSQL QLPLPTTLVQ QQRQLQQQHE 600 EQGDYGEGNM GVVDLSAVAA RVAAFCPDLA PGTDARTLLD SLPLLAQTAF LPHGAGSAAA 660 AALAVGDADA ISLVAAAAPT SVTVSAADSA ASDEGSCSVG VAVGGAVPGE AVKHLVALGR 720 QAQCAATEAM AADAALQAAN QVLEAHQEAA RKAGEVAVSK LGLLKDCIST LAGGGEPLVA 780 GEVGGHFIDL HDLGTVGGLG DMAATGCCYS FDAADLLVAV HS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00048 | PBM | Transfer from AT4G32890 | Download |
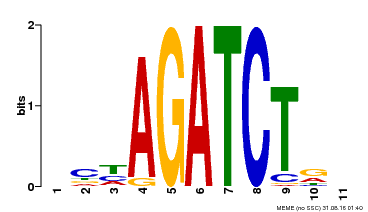
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002957265.1 | 0.0 | hypothetical protein VOLCADRAFT_98336 | ||||
| TrEMBL | D8UF31 | 0.0 | D8UF31_VOLCA; Uncharacterized protein | ||||
| STRING | XP_002957265.1 | 0.0 | (Volvox carteri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32890.1 | 5e-14 | GATA transcription factor 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0003s0248.2.p |



