 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0005s0116.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 1612aa MW: 164626 Da PI: 6.584 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 57.7 | 3e-18 | 1116 | 1166 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
+y+GVr+++ +g+++AeIrdp + r +lg+f+tae+Aa+a+++a+++l+g+
Vocar.0005s0116.1.p 1116 RYRGVRQRP-WGKFAAEIRDPAK---GGRLWLGTFDTAEDAARAYDEAARSLRGA 1166
69*******.**********954...38************************995 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 23.248 | 1116 | 1173 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.9E-11 | 1116 | 1165 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.2E-30 | 1116 | 1173 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.16E-20 | 1116 | 1174 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.04E-28 | 1116 | 1173 | No hit | No description |
| SMART | SM00380 | 2.1E-35 | 1116 | 1179 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-9 | 1117 | 1128 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-9 | 1139 | 1155 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1612 aa Download sequence Send to blast |
MQAPQLSQAA LQLLHPWQPY EVSYKSHHSN VAAAAPPPSA RPAARPVVVI GHQRQGLVQG 60 QGSSGRGLPQ PANGRASSAQ LQQQQQRQQQ RRVSNGDIPV GSTAVAAAAA GSSRRHPQAE 120 TSGDIPVVIN SRGPPSGAAV AAAAAVGVRR QALKATDPPS RTLHLSYQAP QHRLHSMPPA 180 RAQSSQHPHQ THFHHAHHSH PPQPRQPLPP QPPPPQLPYP PRHASAPPSR APQAAPSSSE 240 LHGPSVNTHG RTGNPAMTAS IRGGVLTLQL QLSEIHELAG AADGDNAGYA ITPMDLTSGQ 300 PLGPSLLLST VSLLAAAAAS AAAAAAERAG PLLRESQELP PPLQQQQQQR QQRYHDAGGA 360 ASTAAAAAGS GSYSSQRVQR RSDEESPMEP SSGGNQLQLT PVPEDDKALV DTPAAAPLPL 420 PAPQNGGVGQ LEGTANGVVQ PDDGKGEALR QAARRSGRLG SGGGGGDGDV NSVDSGRDVS 480 GGGGAAHGRS AWDDTAAPAM MSAGNGCGSS GSVAAAAAAA AATALNAYRN GNPAAAAAAL 540 AVFRKYQAQQ QRQRQDRYLD TRMDELDPDK DLYEHERHPS HAKGPGALAR SFDGGGSVRQ 600 DNSDGPDTTV FCNSRHSLDL GAHGAPIGRY DAVRTAAGRY VEAPYDDHDD VDTLATRGFV 660 RRRASEVGPE PSSWSWGGLH EGHRVAAVAA SAEPLRQQQQ MYSQRRLSST YPYRQPPPQC 720 WSYRPVQYVE PPYEGSDNPY GPADEHYEYD KFLPQPTPSA DFLLEGGTRK RPRVASAPAA 780 TAAPGGTAAA AAPSPRSGSG GAAALLDLNA MHVGPRSQNG LSERTTEYVV PEGPHAGVAR 840 LRLPLPVTQL AAAAGRAGVA NASAYMKAAA AAAAAALKSQ NRYGPAAAIG AAADEGKLGA 900 AAGRTDGSAA FPSRHGTATG GDMPSGGAQR AEPLVSPSAR RTDRSAAAGD GATAAAAAAS 960 GHAALHQQAR QKFKAAPERE RGTDMTRAGA AIAAAAMAAV ASAGPGQVAG GGGDDDRDSE 1020 MTESIGVAMS SPGSAAAVAV AGGNGDVDGP VDAAARDGGG GACDAAAAAT GGAAASSPPA 1080 AAAQQPTARG TADAPKGPTP KLKPGPVAAR ARGAVRYRGV RQRPWGKFAA EIRDPAKGGR 1140 LWLGTFDTAE DAARAYDEAA RSLRGAGARV NFPLPAECGE EGEEEGAAAN RALSDGEVHG 1200 EVVQGGGAAA VAGPRGGAGT RGGAASVANG TSGEAAGGAT CWRRTSQNRH GTDPWTGFSG 1260 LEAIVAAAAA VAEEEEAEAR RAAAAAAAAA AAVAESRQPP PQHQEHSPLQ GERRLDVPQR 1320 GPMSVGAVVA AAAPPQRLEA AWRDRGGEFS PLDLDLDHHG DAEGPGAQEH ARQRHHPELQ 1380 QIESSGGGGS GGGRNGAQRS LWSGEDADAK VDSGEGRGEE SGGGPGQEGR VRPVDCPSDG 1440 GGRLVVVPRG VGRALGPARV RTDDLLTAAQ VLLETGGGGR TAFLPLVARL KREPMEYGQP 1500 YAVVRPPTVS LEEEVEVGGH DLAAARMVVA QKMNCPEYSA DETHGSGKQQ QHPQPCTPRE 1560 GDAFPETGID ESVVEVMEEE EEPPQRRQRV DLAEEEVEAA CGEYVDAPIC A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-23 | 1117 | 1180 | 6 | 70 | ATERF1 |
| 3gcc_A | 1e-23 | 1117 | 1180 | 6 | 70 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00548 | DAP | Transfer from AT5G47220 | Download |
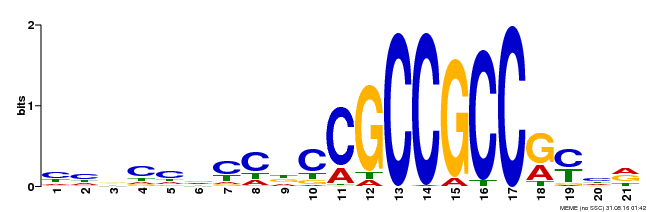
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002951671.1 | 0.0 | AP2 family transcription factor | ||||
| TrEMBL | D8TZ59 | 0.0 | D8TZ59_VOLCA; AP2 family transcription factor | ||||
| STRING | XP_002951671.1 | 0.0 | (Volvox carteri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP132 | 15 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G44210.1 | 3e-12 | erf domain protein 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0005s0116.1.p |



