 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0006s0301.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 3577aa MW: 357162 Da PI: 7.1144 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.8 | 1.4e-14 | 48 | 92 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rW ++E+ ++++a k++G W++I +++g ++t+ q++s+ qky
Vocar.0006s0301.1.p 48 RERWQEDEHARFIEALKLYGRQ-WRKIEEHVG-TKTAVQIRSHAQKY 92
78******************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.33E-13 | 43 | 98 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.867 | 43 | 97 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 3.1E-15 | 46 | 95 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 4.4E-8 | 46 | 96 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.0E-12 | 47 | 95 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-12 | 48 | 92 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.33E-8 | 50 | 93 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009845 | Biological Process | seed germination | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0010243 | Biological Process | response to organonitrogen compound | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043496 | Biological Process | regulation of protein homodimerization activity | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 3577 aa Download sequence Send to blast |
MAQPLLGPSG PERPSSPQPL QQAEVIPAQG AAEAPQKTRK QYTIVKRRER WQEDEHARFI 60 EALKLYGRQW RKIEEHVGTK TAVQIRSHAQ KYFNKIEKAG GSAEAIEVPP PRPKRKSMPA 120 QQRAGAAADT LPTLKFEDNA GSDGGYGPLL GSANGANTYD DDAPLAADLQ QLQLQPLSLP 180 AQAAAHQQQP MQLTQQQLQQ HQLQQQSPQQ GPGVKSYAPQ APPSFRVASS PAAAAAASPG 240 SSGLGLGLGL GLQAPQLPVS MAAAAAAAAA APLAVPPVST QTPATQPTAP QQQQQQQQQL 300 VLPNLLGFPG LGPAAQLQSA FPQPPGGSGA PSAGLTVAPA PAPAPALAPA PAPAPAPQQG 360 QLLGSSLADS IARNIAFNMA QWAQAQVRVA DAPGGLCLGT LGAQQADSSP DRQTVEAVAA 420 AAAAAAAAAA TAVISAAGEA IQQRIQEKAA AGFLPFLVHP LAELIPPPLV ASASAATTSD 480 QSPPLWALPP QQSLMHVLAA SSSGRGRGGG GGGVGRLVSR AHPHSQSQQF TATMLQRTSP 540 FSLDQGAPSG LGGLAGSDVQ THTHTHTHTR GEETTTGQFT GTPGSSQQQQ PAVSEPNQSP 600 QHISPRFAAA AAAAAAAAAA GATGSGSAGL GINSNSNSLM RLASLGPPAS ISLAGTMSLG 660 AGGQGVEQPQ QVLWEGSVPG SAAALSFPVG QAAQNLMRHG SMAVIWDEQQ QQQQQQEQAQ 720 AARDTAALGR LLAGAATFVD QQQQHHQNAA FSFETAQQAA AAAEGLRAAV AVAAIGGNAG 780 GLDASTVAAL SRFGSDTSQQ LQQQQPQRDQ DAAGGLAGLL QQQLLLMQQQ AAAGGPPPVL 840 RSTAPVGQPH QQQLQLTVAS RLAQGSVRGQ LPLPSKQQQQ QLQPSTEGTE TLSPAYQQLL 900 QPQQRLWVEV PRGLDADADA VALQGTPSDP SPASRPPPAA EQCPSPNPAL QQQQQQQQAV 960 ARFSAPAGAA AAVPRPGFAA ESELILNRQH MELEEEEVEE GRGRERARER RSSGRGQGRD 1020 RTSRSDPGGV CSDSAAGGGG GGEPVAAAPS CDKSSGASGC AAGGVDTYQD IEDGRPALQL 1080 LARGGGRQQH HGNPAADITP HFQRQRGSLA GAGAGAALPP RPAGESSPHV EGPEHAATAA 1140 SGDVEGDLLT GKDCLAMRQR LERPHAGDRE PRTGYRLQQR QQEHHRRYAV EREEAEEPEV 1200 LAGGQRHGQP HGASRTTRFK IHMQGPDGAL FDVTTSEKEL HRTHRRLHHH HHYHHHHRHQ 1260 QQTQHLQQQQ EQEQHPPGHR TTRESSQTRQ HQGFQGLVQE QEGEGHTRTH RTRRTAADLD 1320 GELGPTRDLS PHLSHHARGN NARDEAAEAG DGGSGGFGLH PPAANGRERP QQPAAQPPNG 1380 ARLKPHKKER LRPTAPQRPE ALDADAGTAA VATAAARAAA GGASGSWADD FGAATSGDGA 1440 YQLGQLQYLQ HQPGPGLSGT TGSGLNSLQE VLQGPNLEAM VPLLAPATAA AALQQGAALE 1500 QLARLQLNAE HQQHQQHQRR QQQQHQRLQQ QQEQQEHLHL QMPPQGLLLQ QRGGVDGEGA 1560 DVGGPAADAQ DGGSGGNGTD VNAYIRGDYS LMSPSGLQPP HSPSAPSSQQ LPGASGHRNT 1620 PQQQPQPQQQ RTQQPHPQPE QGASQQPQPH RQHRLWACNN NNNNNNQQQQ YSHQNQQLVS 1680 TGHQEMRSSD DRKEATGSGS QLQQPHHHRH HHHHHHNHHH HRHPQHAMQH EQQHSLAQGT 1740 QEEGSSQRSN RPAENRSGNA GGGDASAAAA AAAAGITADG GDAGDASGGA GSGAAGGAAE 1800 VPPAAAAVGA GVVAGTGTVT GTNTGSGSGA GSGAGGGAAD GHQSSGPTGL AVRAAAAHGG 1860 SCMPRQETPP AAVSAGTRQA PPQHQGSSAQ AGTGGTGTGT GGTMCMGMIG ALPPELGSGS 1920 GFGSAGAGGG GGGRQASVGP AGSGSSGADP DPDRKRWEQP GEALPGTSGI GSHVPLHGGL 1980 GPQQQQQHYQ HARARPGTTS VPMAAALKRP WHRAVRGEPA FTGHAAAGST DGEAPADGPD 2040 REEGQGRAAK ERGGAAGGDV HAVRHGDAAG ESVKEHRRGF SGGGGAVAGV IGGSSRHQVE 2100 LMRDAGGQPS CGRQLGAAAS GGAASGGGGG GGGGGGGAAS AGTRKRRSSG SGGAGGSGED 2160 IPAEQPLARQ RPMPMPALTH GLLQLQQAQQ QARAQLQQQH QAVLGAPSSA APPPAHRGGV 2220 DTLALLHSML VQQQQLQVVQ QQQQQQQQPG PSLMSTLAGL SAGAQAVANM LALQQQQQQQ 2280 QQQQQQQQQA HGEAPGQGQG QDVGPPRDVL LASPHQQGMA TITTSNDPLT SSALPAAADA 2340 AAAAAGGTGD GGAEGAADAG HGAPLPDRLQ MAALLSDGGS GGGGCGTSAD PAGGGGLAAL 2400 LPLLSDRERS IESEHIARLE EVARALRYAS SLAGPLAAVL QQPSLQKAFG AGVAAQEVTA 2460 AVNATALQAQ AGDSGNSLPC AAGGPAQPQP EQQQPPPPPQ QQQLQPSSHL AAGADGGGGG 2520 GTLSTAALLQ QWRNAAAAFS LQPLLHDPVP AATAAHTAAA AAAAAMGLGP SGLLPRGQAV 2580 AAGQQSGLSP TLLTYPTHPL LIQQQYMATA QGQADTAAAY LSGLGVGLRA LQPSTTQGLA 2640 VLAGLGGLGL CPSGSAGAGG LLSEQSPRPS LDAAPSAGQT AAAAAVAAPQ PGSQSGLAGP 2700 AAVAAGGRGG LSAGLQHQQQ LEALDGGAGP SGGPAAKPRD PRLNAGNPRT SAGGDGGGAG 2760 GGAGGGLFRH PTNPLTGTGD GAVGAAGPTK SFGRVDGGVS GGGRNNLGYV PESGGSGGDR 2820 AVGKVGQGAG GVVASGKRNV SELYGNHAAA AATAAGATQR VGAAAPRHKP SRRRHHRGHS 2880 PQRSVGSGAS SSGFHCGGDK SGGEEANHCP RPSREYCNKD GLAPGGGLGT EQQDTATSSG 2940 KDGEDGAGAG GVMAGGNSAG AGDAAGAAGA MPPSKLPRIG HEAPGKLQLH GGGGCGAAGA 3000 APLPVGRTAH GHHVPTEATP SSGGGSGGSG GGSIGGNKSG TSNDAYDGSM NASDEGSNEP 3060 GTAPADGPMG QPMAPSDGEG RLGMGRDGDA LNVQGMGGAG GGGMICGGAR SFSVAQPDPP 3120 SFGQAMATVQ GYMVAEHAGA AAVPSGVGHK RQRAEMQNGG GGGAAVTAAT AAMAAARQPD 3180 AGSGGGGAAG GGGGMLPQPP HKAPRTASSS GPHGPVPGAN PASAAAAGGA GLAQPCRGGG 3240 GGGDGSNADN GSHGATEGED GEVGGSGNRN RTPPGALREQ QSARAVGGTP PAQQQPAGSL 3300 PSHLAYILEQ GSSKQGGNDT GHNNSGTGPR SGVGSNEGGH GGNGGSRSNE GGNGNGSQGN 3360 GSHGNGQGSN GNGSHGHGSN GNNDNGSNGN GASTNLPGSG RHTYCRTQAG LAHRGGGTRL 3420 TGGGSGGSGG SGGSGGGGGA RALDGASGGG LLSPYDDTPV AGRTSQPPHH HHHHHHHHVD 3480 TALEGTSMRT QPPVPQGDMD PEPNQRLPQW QQQPQPQQPQ PQCGMQTGSA QAMEAILVNV 3540 GDGVWDAADV AGRRAPLGGA TACAGADVTG ALPEGF* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2142 | 2147 | TRKRRS |
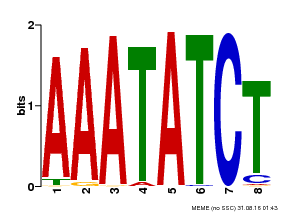
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00103 | PBM | Transfer from AT2G46830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP454 | 14 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02840.3 | 1e-29 | LHY/CCA1-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0006s0301.1.p |



