 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0014s0006.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 846aa MW: 84658 Da PI: 8.0639 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 20 | 1.2e-06 | 246 | 265 | 6 | 26 |
-SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 6 CrffartGtCkyGdrCkFaHg 26
C+++ + G+C +Gd C++aHg
Vocar.0014s0006.1.p 246 CPDYKQ-GFCIRGDVCPYAHG 265
******.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.25.40.20 | 3.6E-8 | 81 | 136 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.02E-7 | 81 | 135 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.084 | 82 | 114 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 11.851 | 82 | 148 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.55E-5 | 84 | 135 | No hit | No description |
| Pfam | PF13857 | 7.2E-7 | 85 | 123 | No hit | No description |
| PROSITE profile | PS50103 | 11.967 | 240 | 267 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 0.0018 | 240 | 266 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 2.8E-4 | 246 | 265 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 1.9E-4 | 272 | 304 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 6.822 | 275 | 299 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 16 | 275 | 298 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 846 aa Download sequence Send to blast |
MESEVIGHYL IASNGGPATP LKNESTVLFQ DVVSDKLSNV EAAIEADASK LHQFFLILHW 60 SGDANGALDA NGAAAKTSVM VKQRTLLMIA AHAGSLRVLA YLLARGAEPS RKSADGLTAY 120 DLAVAGESSQ AATALAMMRD ADSHPSSSTR SSSTTETQSF TGTDAVRGVN RQGAQDSVAD 180 SEGSSYSTTD LTRPEYSTDD FRMFNFKVLR CSKRHAHDWR ACPFAHPTEN ARRRDPREFK 240 YCALACPDYK QGFCIRGDVC PYAHGVFECW LHPSRYRTQL CKDGANCHRP VCFFAHSLPE 300 LRAPTYTWVP SPADLAKPPA AAATAASSGT SSALQSSTSM TAASAMALGG GADGAVAGGP 360 PPVQVGSPMA VTGPEGRSLL STASSGLPPA STSSDGTSSG IGPSCGSPRT LNGSGSGMLQ 420 QLQLGMGPGS GLVGSGCGGM VMMTSACNGN GNGLAVGTSS SSCSSGANGS PSPSCSIGSK 480 DGGCANGNGN GNGVVAGGIL AVVPAATANG GGGGKEGKGS SGAGGTPGSG GKGNQNLTQP 540 NAAASMFGFT APRMSNAFAR RHGLNPKDNP MLNLQKIALQ SQQQQQQQQQ QQVQQVQQVQ 600 QQQQQAQNSA LGIAGGHLGA FGPQDGAAAL RNHAAVAAAG NGAGIIQAAA AAAGTAGSRA 660 AANFNSSANG QRLQVAAAAA AAAAAAAANS ATSPGAFSTV PFANPAASPF GNHVAPLHAH 720 MQMHAAAAAA AAHLGANNPA DPSLLAAQLA ALNIQQQQQQ QQQQQRQQHL GIGIGIGGIN 780 MASGAAAAAA AAAAAGIGHG PVQYVASAAP HPALIHTGMY GPGGGMYDTA QQHLMVAMGG 840 AQAEV* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00566 | DAP | Transfer from AT5G58620 | Download |
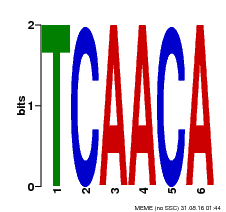
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002958507.1 | 0.0 | hypothetical protein VOLCADRAFT_99797 | ||||
| TrEMBL | D8UIN8 | 0.0 | D8UIN8_VOLCA; Uncharacterized protein | ||||
| STRING | XP_002958507.1 | 0.0 | (Volvox carteri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP60 | 15 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40140.2 | 5e-50 | C3H family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0014s0006.1.p |



