 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vocar.0019s0209.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Volvox
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 415aa MW: 43153.1 Da PI: 5.0877 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 36.5 | 8.7e-12 | 127 | 174 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
++ rE+ RR+++N++f L +l+ + + +K +K++iL+ A++Y+++
Vocar.0019s0209.1.p 127 KKACREKARREKLNERFLDLARLVDPG---NEPKTDKSTILTDAIKYVQQI 174
5678**********************8...9*****************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 13.535 | 123 | 174 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.53E-9 | 126 | 173 | No hit | No description |
| SuperFamily | SSF47459 | 7.07E-14 | 126 | 190 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.5E-9 | 127 | 173 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.8E-11 | 129 | 180 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.9E-12 | 130 | 188 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MEAPCSLEQS CKLEQAGKGG AADDALNSSF LEFLTDDLVF GSEALDSTTL QAICQVADAQ 60 RELAGVSGAA VSAATQPGPL AASAQGMKRP LDDAGFSESD EEDEAGRGEK SAGKRTKGGE 120 MSAAATKKAC REKARREKLN ERFLDLARLV DPGNEPKTDK STILTDAIKY VQQITVENHQ 180 LKQLNKFLEV RRGNSKPGLS ACMFPEVGWS LTLFSTLEQE RVSSLERERG QQLYQHSLMM 240 MGQGQMMPAG QAMLVGGPAG MMACPVIAGA PMGHPHLQMP TAMAASAGNP LLSAASLPSA 300 STPPLPTMSN QQQQAGQASV LAQAQGGLLA QAQGAMMTQM PLPAATGTAL GPVGASVSTA 360 GVSFNVAGAN AGAGPSKPIM VNTVQGMMPY QGMYWLPPQM MDSTQDSLLR PPAA* |
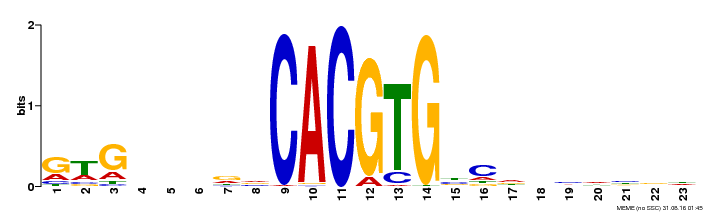
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00372 | DAP | Transfer from AT3G23210 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2J8A8V8 | 2e-91 | A0A2J8A8V8_9CHLO; Transcription factor bHLH34 | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP5649 | 7 | 7 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23210.1 | 7e-11 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Vocar.0019s0209.1.p |



