 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vradi0086s00350.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 255aa MW: 28959.5 Da PI: 9.3331 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 125.9 | 1.6e-39 | 128 | 205 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cq+e+C+adl eak+yhrrhkvCe h++a+vvlv+g++qrfCqqCsrfhelsefD++krsCrrrLa hnerrrk+++
Vradi0086s00350.1 128 CCQAEKCNADLHEAKQYHRRHKVCESHAEAQVVLVHGTKQRFCQQCSRFHELSEFDDAKRSCRRRLAVHNERRRKNSS 205
6**************************************************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.7E-31 | 126 | 190 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 30.983 | 126 | 203 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.36E-36 | 127 | 206 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.4E-31 | 129 | 202 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MNGGASYSEE AHSGRDEARR NLRQRLSADM LRLNEPHKNI IVYSPVTTVT IPPLYHNSHT 60 PSPLFNDSTC HSSMEGLEPK VVVRGERLKR DVDCGDNQKG YVELRQENKK KKKGEGGKKG 120 CVDGGMLCCQ AEKCNADLHE AKQYHRRHKV CESHAEAQVV LVHGTKQRFC QQCSRFHELS 180 EFDDAKRSCR RRLAVHNERR RKNSSDQSQA EGSRHHKGSE VPQMKDIACG QANERGRTHI 240 TIQENSAYKK FEIR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 8e-33 | 125 | 202 | 7 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 108 | 118 | KKKKKGEGGKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Promotes both vegetative phase change and flowering. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16914499}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00634 | PBM | Transfer from PK22320.1 | Download |
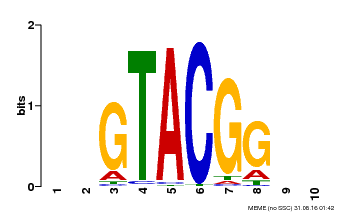
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vradi0086s00350.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156 and miR157. {ECO:0000305|PubMed:12202040, ECO:0000305|PubMed:16914499}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015040 | 1e-149 | AP015040.1 Vigna angularis var. angularis DNA, chromosome 7, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022633611.1 | 1e-130 | squamosa promoter-binding-like protein 3 | ||||
| Swissprot | Q9S7A9 | 3e-38 | SPL4_ARATH; Squamosa promoter-binding-like protein 4 | ||||
| TrEMBL | A0A1S3TD82 | 1e-129 | A0A1S3TD82_VIGRR; squamosa promoter-binding-like protein 3 | ||||
| STRING | XP_007149828.1 | 9e-85 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF535 | 34 | 153 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53160.2 | 5e-40 | squamosa promoter binding protein-like 4 | ||||



