 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vradi0221s00140.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 312aa MW: 34994.9 Da PI: 6.5131 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.6 | 2.4e-32 | 64 | 117 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+av++LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
Vradi0221s00140.1 64 PRLRWTPDLHLRFVHAVQRLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 117
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-29 | 60 | 118 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.929 | 60 | 120 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.32E-15 | 63 | 118 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.4E-23 | 64 | 119 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.1E-9 | 65 | 116 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MEEERHVSEC SKTSPSIEEG CEESEEENED ESKQNNNGGS SSNSTVEENE KKATVRPYVR 60 SKLPRLRWTP DLHLRFVHAV QRLGGQERAT PKLVLQLMNI KGLSIAHVKS HLQMYRSKKV 120 EANQGRIRYG YGDASLAIYE NMVKTCMNRS SVDESGAEFC GSRLTERANG TNSNIIWNHN 180 VLQADCSGFN ESFCRPIKMN QEDPHQSTEM QPSAQELMPN NDMPSKNLVE LKSLKRKASD 240 SDIDLNLSLK LSSRVSDENH GSMVEHEIDS NLSLSLCSQS SSSYLSSRLR KSSKDHSKEQ 300 GERGRTLDLT I* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 1e-17 | 65 | 119 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-17 | 65 | 119 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-17 | 65 | 119 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-17 | 65 | 119 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-17 | 65 | 119 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
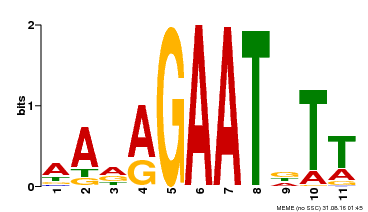
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vradi0221s00140.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015037 | 0.0 | AP015037.1 Vigna angularis var. angularis DNA, chromosome 4, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014490662.1 | 0.0 | two-component response regulator ORR22 | ||||
| TrEMBL | A0A1S3TA67 | 0.0 | A0A1S3TA67_VIGRR; two-component response regulator ORR22 | ||||
| STRING | XP_007140226.1 | 1e-141 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 1e-42 | G2-like family protein | ||||



