 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vradi05g00550.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 935aa MW: 106100 Da PI: 7.1299 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 149.6 | 7.8e-47 | 92 | 213 | 3 | 117 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
+ k+rwl+++ei+aiL n++ +++++++ + p+sg+++L++rk++r+frkDG++wkkk dgktv+E+he+LKvg+ e +++yYah+++n+tf r
Vradi05g00550.1 92 EAKSRWLRPNEIHAILCNYKYFKIKAKPVHLPESGTIVLFDRKMLRNFRKDGHNWKKKSDGKTVKEAHEHLKVGNEERIHVYYAHGQDNSTFVR 185
459******************************************************************************************* PP
CG-1 97 rcywlLee.......elekivlvhylev 117
rcywlL++ +le+ivlvhy+e+
Vradi05g00550.1 186 RCYWLLDKyvlldfrNLEHIVLVHYRET 213
******974444333579*******997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 70.421 | 86 | 219 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-67 | 90 | 214 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.1E-42 | 92 | 212 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 2.5E-6 | 359 | 473 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 5.25E-16 | 384 | 471 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 1.71E-17 | 561 | 682 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 3.4E-7 | 562 | 649 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.21E-14 | 569 | 679 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.2E-15 | 574 | 683 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.539 | 587 | 652 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.007 | 620 | 652 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3.3E-5 | 620 | 649 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2300 | 659 | 690 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.96E-5 | 750 | 831 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 83 | 783 | 805 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.474 | 784 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 5.0E-4 | 806 | 828 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.127 | 807 | 831 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.9E-4 | 808 | 828 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 4.96E-5 | 883 | 910 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 11 | 883 | 905 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.078 | 885 | 913 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 935 aa Download sequence Send to blast |
KYEKGTEESS VIIIQLPAAF FFLPILTYPS SNQVVGDAIV GFVVREGGVG GENLHLVCLF 60 LMAHDLTGQL VGSEIHGFHT LQDLDVSNTM EEAKSRWLRP NEIHAILCNY KYFKIKAKPV 120 HLPESGTIVL FDRKMLRNFR KDGHNWKKKS DGKTVKEAHE HLKVGNEERI HVYYAHGQDN 180 STFVRRCYWL LDKYVLLDFR NLEHIVLVHY RETREIQLQS SPVTPVNSNS SSVSDPAASW 240 ILAEDLDSGA KSAHSAELND NLTIKSYEEK LHEINTLEWD DLVVSNVNTS TTSNGGNVPH 300 SYQQNQSLLS GSFDNVSVTP LAEVPPLGLQ SQDSFGTWMN NIITDTPCSV DESALETSIS 360 IVREPYSSLV ADNQLSSLPE QVFNLTEVSP AWASSTEKTK VLITGYFHSN YQHLAKFNLV 420 CVCGDVSFPV EIVQVGVYRC WVPPHSPGLV NIYLSFDGHK PISHVVNFEY RTPILHDPTA 480 TMEEKYNWNE FRLQMRLAHL LFSTDKILDI FSNTVSPNAL KEASRFSFRT SFISKSWQFL 540 MKSIDEQTTP FPQVKDSLFE IALKNKLKEW LLERIVVGSK STEYDAQGQG VLHLCAMLGY 600 SWAISLFSWS GLSLDFRDKF GWTALHWAAY YGMEKMVATL LSAGARPNLV TDPTPKNPGG 660 FTAADLAYMK GYDGLAAYLS EKSLVEQFND MSLAGNISGT LDTSSTDVVN AQELTEDQLY 720 LKETLAAYRT AAGAAARIQA AYKEHELNLR YKAVELNTPE HQARRIVAAM KIQHAFRKYE 780 TRKSNAAAVR IQLRFRTRKI RREFLNMRRQ AIKIQAAFRG FQVRRQYKKI IWSVGVLEKA 840 ILRWRLKRKG FRGLQVNSVE DGEQEDGVEE EFFRTGLKQA EERVERSVVR VQAMFRSKKA 900 QEEYRRMKLA YSQAKLDLEF EDFLSSEVDM LTDI* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
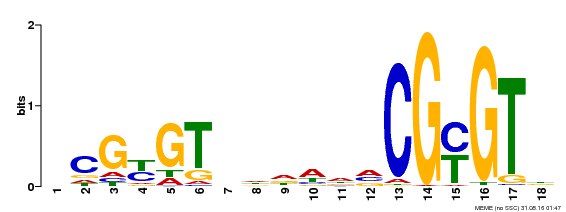
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vradi05g00550.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015035 | 0.0 | AP015035.1 Vigna angularis var. angularis DNA, chromosome 2, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014499049.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A1S3TZ74 | 0.0 | A0A1S3TZ74_VIGRR; calmodulin-binding transcription activator 5 | ||||
| STRING | XP_007138978.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16940.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



