 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vradi07g29890.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 876aa MW: 97665.8 Da PI: 6.8635 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 75.7 | 5.3e-24 | 159 | 260 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++ +++a+e+ + k++ +++l+ +d++g++W++++i+r++++r++l++GW+ Fv++++L +gD ++F + +
Vradi07g29890.1 159 FCKTLTASDTSTHGGFSVLRRHADEClppldmS-KQPPTQELVAKDLHGNEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGE 249
99*****************************63.4445569************************************************..449 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
++el+v+v+r+
Vradi07g29890.1 250 NGELRVGVRRA 260
999*****996 PP
| |||||||
| 2 | Auxin_resp | 100.3 | 2.6e-33 | 285 | 377 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnP..........rastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+ t+++F+v+Y+P r+s++eF+v++++++++lk+++++GmRfkm+fe+e+++e+r++Gt+vg++d+d+ rWpnSkWrsLk
Vradi07g29890.1 285 AWHAILTGTMFTVYYKPsiysnvqiiyRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGTIVGIEDADQKRWPNSKWRSLK 377
79***************77777777777899************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 1.1E-41 | 155 | 274 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.31E-40 | 155 | 288 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.08E-21 | 157 | 259 | No hit | No description |
| PROSITE profile | PS50863 | 12.026 | 159 | 261 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.9E-22 | 159 | 260 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.8E-23 | 159 | 261 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.5E-31 | 285 | 377 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 24.592 | 742 | 835 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 5.89E-11 | 743 | 819 | No hit | No description |
| Pfam | PF02309 | 2.7E-8 | 788 | 830 | IPR033389 | AUX/IAA domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 876 aa Download sequence Send to blast |
MASSEVSIKG NSVNGKGENS SGGFNGCNDV RNAGGELQNA SSSSSARDAE TALYRELWHA 60 CAGPLVTVPR EGERVFYFPQ GHIEQVEAST NQVAEQHMPV YDLPPKILCR VINVMLKAEP 120 DTDEVFAQVT LLPEPNQDEN AVEKEGPPAP PPRFHVHSFC KTLTASDTST HGGFSVLRRH 180 ADECLPPLDM SKQPPTQELV AKDLHGNEWR FRHIFRGQPR RHLLQSGWSV FVSSKRLVAG 240 DAFIFLRGEN GELRVGVRRA MRQQGNVPSS VISSHSMHLG VLATAWHAIL TGTMFTVYYK 300 PSIYSNVQII YRTSPAEFIV PYDQYMESLK NNYTIGMRFK MRFEGEEAPE QRFTGTIVGI 360 EDADQKRWPN SKWRSLKVRW DETSNVPRPE RVSQWKIEPA LAPPALNPLP MPRPKRPRSN 420 VVPSSPDSSV LTREASSKVS VDPLPASGFQ RVLQGQELST LRVNFAESNE SDTAEKSAWS 480 SAADDEKIDV VSTSRRYGSE SWMSMGRHEP TYPDLLSGFG VHGDQTSHPS LVDQNGPIAN 540 LSRKHYFDRE GKHNVLSPWP VVPSSLSLNL LDSNTKVSAQ GGGDTTSQVR GNLRFSSAFG 600 DYTVLHGHKV DHSHGNFLMP PPLSTQYESP RSRELLPKPI SGKPSEVSKP KDSDCKLFGI 660 SLLSSPIALD PSVSQRNVAI EPVGHMHTIH QQRTYENDPK FENSRGLKPA DGVLIDDHEK 720 PTQTSQPHLK DVQPKSNSGS ARSCTKVHKK GIALGRSVDL TKFSAYDELI AELDELFEFG 780 GELTSPQKDW LIVYTDNEGD MMLVGDDPWQ EFVAMVRKIY IYPKEEIQKM SPAESFSFRG 840 LIIRIDFDFH SFQIPNWSEW KVVGDCHKAI PSWRI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-174 | 52 | 399 | 18 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-174 | 52 | 399 | 18 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-174 | 52 | 399 | 18 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
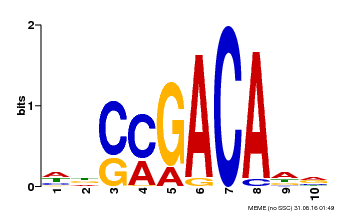
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vradi07g29890.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 0.0 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014508669.1 | 0.0 | auxin response factor 2B | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | A0A1S3URW9 | 0.0 | A0A1S3URW9_VIGRR; Auxin response factor | ||||
| STRING | XP_007159966.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3281 | 33 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||



