 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Vradi08g11790.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Vigna
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 764aa MW: 82347.4 Da PI: 6.2988 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.5e-16 | 461 | 507 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Vradi08g11790.1 461 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 507
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.36E-20 | 454 | 517 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.8E-20 | 454 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 457 | 506 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.88E-17 | 460 | 511 | No hit | No description |
| Pfam | PF00010 | 1.2E-13 | 461 | 507 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 463 | 512 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 764 aa Download sequence Send to blast |
LAREKLGTEN NSTRASDQCS SPENDFFELV WENGQISSQG QSSKVRRSPS CRSLPSHCLP 60 SHSPKGRDKD VGYGANRMGK FRDLDSGLNE ISMSVPSREV DFCQDEDVLP WFDYPTMDDS 120 LQHDYGSDFL PAPTSFTLSD KKSNSSMALR DSHKTSEEQG NAFDDSSAEQ VGNVEPKAIT 180 NQLYPPSLHH CQTSFVSSRS RTSDITENNS TSNANQDATY GEITNFLSSS SDFSSPKGKK 240 QEPPLPGNGS SSTIMNFSHF ARPAAKVRAN LQNIGLKSGL SSARSDSMEI KNKDAAATSS 300 NPPESVVVDS GGECPKELTI HCQQNVDQSK TNLNTSLSKA VEQSAVLSKQ SETAFNESST 360 KIDKIADRVL GDSGVKGQTT AEKSMEAAVA SSSVCSGNGA DRGSDEPNKN VKRKRKDTDD 420 SECHSERTFY MKDAEEESAG VKKAAGGRGG TGSKRTRAAE VHNLSERRRR DRINEKMRAL 480 QELIPNCNKV DKASMLDEAI EYLKTLQLQV QIMSMGAGLY MPPMMLPAGM QHMHAPHMTP 540 FSPLGVGMHM GYGMGYGVGM PDMNGGSSRF PMIQVPQMQG SHIPVAQMSG PTALHGMTGS 600 NLPRLGLPGQ GHPMPMPRAP VLPFSGGPVM NSSALGLNAC GLSGLVETVD SASASGLKDQ 660 MPNVGPLVKQ SIGGGDSTSQ MPTQSIAIPM INFHFNENIL LPFFYSSLHV CVLQFCYARV 720 KQLLLDLSNL LWFITAVIPL GLMTMELLIL VPADNVMSGY DPE* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 465 | 470 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
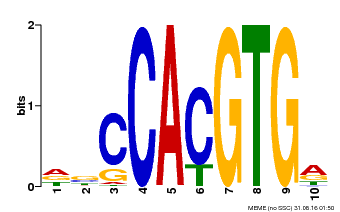
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Vradi08g11790.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015041 | 0.0 | AP015041.1 Vigna angularis var. angularis DNA, chromosome 8, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022640840.1 | 0.0 | transcription factor PIF3 isoform X1 | ||||
| TrEMBL | A0A3Q0F9U6 | 0.0 | A0A3Q0F9U6_VIGRR; transcription factor PIF3 isoform X1 | ||||
| STRING | XP_007145054.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5271 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-51 | phytochrome interacting factor 3 | ||||



