 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00001657-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 319aa MW: 36262.3 Da PI: 4.319 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.3 | 1.3e-18 | 87 | 140 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t +q++ Le+ Fe ++++ e++ +LAk+lgL+ rqV +WFqNrRa+ k
WALNUT_00001657-RA 87 KKRRLTVDQVQLLERNFEVENKLEPERKVQLAKELGLQPRQVAIWFQNRRARFK 140
56689***********************************************88 PP
| |||||||
| 2 | HD-ZIP_I/II | 126.5 | 1.2e-40 | 87 | 177 | 2 | 92 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
kkrrl+ +qv+lLE++Fe e+kLeperKv+la+eLglqprqva+WFqnrRAR+k kqlEkdy++Lk++yd+lk + ++L +e+e+L++e+
WALNUT_00001657-RA 87 KKRRLTVDQVQLLERNFEVENKLEPERKVQLAKELGLQPRQVAIWFQNRRARFKIKQLEKDYDSLKASYDRLKVDYDNLLEEKESLKNEVI 177
9**************************************************************************************9985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.11E-18 | 73 | 143 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.827 | 82 | 142 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.2E-17 | 84 | 146 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.30E-17 | 85 | 143 | No hit | No description |
| Pfam | PF00046 | 5.9E-16 | 87 | 140 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 6.1E-19 | 89 | 150 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.9E-6 | 113 | 122 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 117 | 140 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.9E-6 | 122 | 138 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.6E-16 | 142 | 182 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MVGGEVYGSA SNMSVLLQNE RLPCSSEVLE PLWIPGGPNP SLHASKSMVN YEDVNGGDAA 60 ARPFFQPLDK DESVDDDYDG CFHQPGKKRR LTVDQVQLLE RNFEVENKLE PERKVQLAKE 120 LGLQPRQVAI WFQNRRARFK IKQLEKDYDS LKASYDRLKV DYDNLLEEKE SLKNEVISLE 180 EKFLVREKVK ENVEPLDATK LSSAETQEPI PNAVSEIMSI VAIPVCKQED ASSAKSDVFD 240 SDSPHYTDGN HSSLLEPAYS SHVFEPDQSD FSQDEEDDRS KIPLPQAFYL KVEDGCNEPA 300 NYCNFGFPVE DQPLLYWNY |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 134 | 142 | RRARFKIKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
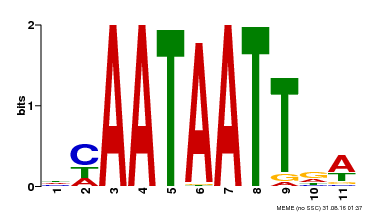
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018831127.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HAT5-like | ||||
| TrEMBL | A0A2I4FHL1 | 0.0 | A0A2I4FHL1_JUGRE; homeobox-leucine zipper protein HAT5-like | ||||
| STRING | XP_007157405.1 | 1e-135 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1500 | 34 | 99 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 7e-42 | homeobox 1 | ||||



