 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00003701-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 299aa MW: 33595.6 Da PI: 8.163 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.2 | 6.7e-19 | 135 | 189 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ +++k+q +Le+ F+++++++ +++ LAk+lgL rqV vWFqNrRa+ k
WALNUT_00003701-RA 135 RKKLRLSKDQSAILEDSFKEHNTLNPKQKLALAKQLGLRPRQVEVWFQNRRARTK 189
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.8 | 1.1e-41 | 135 | 224 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+kk+rlsk+q+++LE+sF+e+++L+p++K +la++Lgl+prqv+vWFqnrRARtk+kq+E+d+e+Lkr++++l++en+rL+kev+eLr +l
WALNUT_00003701-RA 135 RKKLRLSKDQSAILEDSFKEHNTLNPKQKLALAKQLGLRPRQVEVWFQNRRARTKLKQTEVDCEFLKRCCENLTDENRRLQKEVQELR-AL 224
69*************************************************************************************9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04618 | 5.2E-34 | 1 | 109 | IPR006712 | HD-ZIP protein, N-terminal |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-18 | 122 | 185 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.24E-18 | 127 | 192 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.346 | 131 | 191 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 7.2E-17 | 133 | 195 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.8E-16 | 135 | 189 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.86E-15 | 135 | 192 | No hit | No description |
| PRINTS | PR00031 | 2.0E-5 | 162 | 171 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 166 | 189 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 2.0E-5 | 171 | 187 | IPR000047 | Helix-turn-helix motif |
| SMART | SM00340 | 2.9E-27 | 191 | 234 | IPR003106 | Leucine zipper, homeobox-associated |
| Pfam | PF02183 | 2.4E-11 | 191 | 225 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009641 | Biological Process | shade avoidance | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 299 aa Download sequence Send to blast |
MVEKEDLGLS LSLSFPQNNR SLQLNLRPSL LPSSAASPSG FNLPKPSWNE AAFPSSDRNS 60 ESCRAETRSF LRGIDVNRLP SSGDCEEEAG VSSPNSTISS LSGKRSEREA NGEELEIERD 120 CSRGASDEED GETSRKKLRL SKDQSAILED SFKEHNTLNP KQKLALAKQL GLRPRQVEVW 180 FQNRRARTKL KQTEVDCEFL KRCCENLTDE NRRLQKEVQE LRALKLSPQF YMQMTPPTTL 240 TMCPSCERVA VPVCSSATVD TRPHHQVVST HARHVSINPW ATTPGPIARR PFDSHHQRS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 133 | 139 | SRKKLRL |
| 2 | 183 | 191 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that plays a role in auxin-mediated morphogenesis. Negatively regulates lateral root elongation. {ECO:0000269|PubMed:12492842}. | |||||
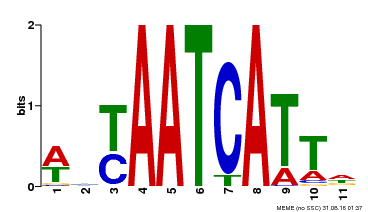
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00550 | DAP | Transfer from AT5G47370 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By indole-3-acetic acid (IAA). {ECO:0000269|PubMed:12492842}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018839633.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HAT4-like | ||||
| Swissprot | P46601 | 1e-109 | HAT2_ARATH; Homeobox-leucine zipper protein HAT2 | ||||
| TrEMBL | A0A2I4G6W7 | 0.0 | A0A2I4G6W7_JUGRE; homeobox-leucine zipper protein HAT4-like | ||||
| STRING | EOY24263 | 1e-163 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1239 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47370.1 | 1e-95 | Homeobox-leucine zipper protein 4 (HB-4) / HD-ZIP protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



