 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00007052-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 398aa MW: 44931.1 Da PI: 9.1004 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.1 | 6.1e-11 | 112 | 153 | 3 | 44 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNka 44
++k+ rr+++NReAAr+sR+RKka++++Le+ +L + ++
WALNUT_00007052-RA 112 ADKVKRRLAQNREAARKSRLRKKAYVQQLETSRLKLAQLEQE 153
79****************************854444444333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.4E-4 | 110 | 168 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 7.7E-8 | 110 | 159 | No hit | No description |
| Pfam | PF00170 | 8.0E-8 | 112 | 155 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.117 | 112 | 156 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.37E-6 | 114 | 155 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 117 | 132 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 3.0E-30 | 200 | 275 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 398 aa Download sequence Send to blast |
MVEMGEMPFH RLMMSSKAVM DSCATLWWAH VKRRCAKVMS SASIQPFTSR RMGIYNPFQQ 60 VSLWGDTFKG DSSPNTGAST VLQVDAGMHN KSEIISQESM GPSRNDEGSN SADKVKRRLA 120 QNREAARKSR LRKKAYVQQL ETSRLKLAQL EQELEKARKQ GMYGQSALDT SHGGFSGNVN 180 SGIAAFEMEY AHWVEEQNRQ ICELRSALQA HITEIELRML VESGLNHYHS LFRMKADAAK 240 ADVFYLMSGV WRTSTERFFL WIGGFRPSEL LNIIRPQLEP MTDQQVLNVC NLRQSSQQAE 300 DALSQGMEKL QQSLAQSIGI ESVIAGNYGS QMAAEMEKLE ALEGFVNQAD HLRHQTLQQM 360 SRILTTHQTA RGLLALGEYF HRLRALSSLW AARPREPA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
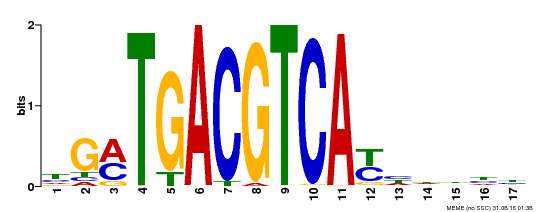
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018849635.1 | 0.0 | PREDICTED: transcription factor TGA7-like | ||||
| Swissprot | Q93ZE2 | 1e-169 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A2I4H0F2 | 0.0 | A0A2I4H0F2_JUGRE; transcription factor TGA7-like | ||||
| STRING | VIT_18s0001g04470.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1096 | 34 | 104 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 1e-152 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



