 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00010075-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 530aa MW: 59031.8 Da PI: 6.3789 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21 | 8.9e-07 | 269 | 290 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
C++Cgk F+r nL+ H+r H
WALNUT_00010075-RA 269 FCTICGKGFKRDANLRMHMRGH 290
6*******************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.25E-5 | 266 | 293 | No hit | No description |
| SMART | SM00355 | 0.0026 | 268 | 290 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.03 | 268 | 295 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.9E-6 | 269 | 319 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 270 | 290 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 53 | 317 | 350 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 22 | 355 | 377 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010044 | Biological Process | response to aluminum ion | ||||
| GO:0010447 | Biological Process | response to acidic pH | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 530 aa Download sequence Send to blast |
MDHKENMWTK SFSSGNELLK NISTGQPSFP NFDSQQSQEK WEDPSMLDYS TRIEPPIKEF 60 KQHSEAQSSL PCNPNNQVKL PVQEDGQMNG SLQSSKIQDW DPRAMLNNLS FLEQKIHQLQ 120 DLVHLIVGRR GQVFERPDEL AAQQQQLITA DLTSIIVQLI STAGSLLPSV KHNTLSSTTT 180 VGQLGGVYLP SMVDLNGGFQ VQDNGESKVS DQSNPIDLAG NIGNEQIYHI EEHESKDEED 240 ADEGENLPPG SYEILQLEKE EILAPHTHFC TICGKGFKRD ANLRMHMRGH GDEYKTPAAL 300 AKPNKESSSE PILLKRYSCP YAGCKRNKDH KKFQPLKTIL CVKNHYKRTH CDKSYTCSRC 360 NAKKFSVIAD LKTHEKHCGK DKWLCSCGTT FSRKDKLFGH IALFQGHTPA ISIDEIKGQS 420 GPPDRGEGNE SINKVRSINF NFSSGAPSRS EVQNAVDVKA GIEDPASYFS PLNFDTCNFG 480 GFHEFPRPPF EDSESSFSFF IPGSSSNYTV PGSCNYTKKT GGESSSNNLE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP2, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:17535918, ECO:0000269|PubMed:18826429, ECO:0000269|PubMed:19321711, ECO:0000269|PubMed:23935008}. | |||||
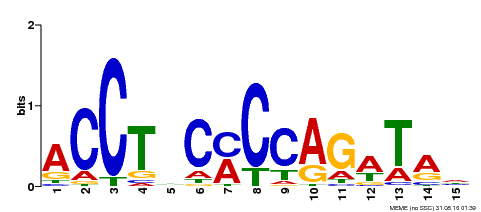
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00196 | ampDAP | Transfer from AT1G34370 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By shock H(+) and Al(3+) treatments. {ECO:0000269|PubMed:17535918}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018825336.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Refseq | XP_018825337.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Refseq | XP_018825338.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| Swissprot | Q9C8N5 | 0.0 | STOP1_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| TrEMBL | A0A2I4F109 | 0.0 | A0A2I4F109_JUGRE; protein SENSITIVE TO PROTON RHIZOTOXICITY 1 isoform X1 | ||||
| STRING | XP_008237278.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5340 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34370.2 | 0.0 | C2H2 family protein | ||||



