 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00012788-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 838aa MW: 92027.3 Da PI: 6.7625 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.7 | 7.4e-16 | 545 | 583 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
WALNUT_00012788-RA 545 SCKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQECFNKP 583
689**********************************96 PP
| |||||||
| 2 | TCR | 50.9 | 3.2e-16 | 630 | 668 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
WALNUT_00012788-RA 630 RHKRGCNCKKSNCLKKYCECYQGGVGCSISCRCEGCKNA 668
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 2.5E-17 | 544 | 585 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.543 | 545 | 670 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.7E-11 | 547 | 582 | IPR005172 | CRC domain |
| SMART | SM01114 | 5.6E-17 | 630 | 671 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.2E-11 | 632 | 668 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 838 aa Download sequence Send to blast |
MDTPERNQIG TPISKFEFWT NFVHVYLAVF MESSDVYARN VVYHCKQDSP VFNYINSLSP 60 IKPVKSIHIT QTFNPLNFAS LPSVFTSPHV NSHKESRFLK RHNCLDPSKP ESSSGNGNKI 120 GTNEGTGVDA AQLHDNSAEL QENFDSEVSL KVASAQPSSE HSKFVIELPC TLKYDCGSPE 180 CGVTLSCDFE IGCMSELVSK SASLIQYGQE ASGNGSSEGE VHLSEMCRIG QKKEGMECDW 240 ESLISDATDL LIFNSPNNSE AFKGSIQKST DPRIRFSTSL MSELHQNDIK NLQTMQIVDP 300 IGSGEQYETD NPSAETGEGN ELQEIEQTQD NAAESNLSKG MSSHMREQVE SEVAECIPYA 360 SKAVSNLHRG MRRRCLDFEM AGARRKALGD ASNFSSSMLY EKINCYDKQL VAIKPSGDSS 420 RCILPGIGLH LNALATTSKD QKNVKHESLS SGIQLYLPSS TASLHSPTNS QEPLHKSLTS 480 VSAERDTDLA ENGVSHVEDA SQASAYLVSE ELNQNSPKKK RHVHLCIYLN WIFRRRLEHA 540 GESESCKRCN CKKSKCLKLY CECFAAGVYC IEPCSCQECF NKPIHEDTVL ATRKQIESRN 600 PLAFAPKVIR SSESVTEIGD ESSKTPASAR HKRGCNCKKS NCLKKYCECY QGGVGCSISC 660 RCEGCKNAFG RKDGCAPIGT EADIEEETDA CEMEKAIQKS DIQNNEEQNP ATTLPTTPLR 720 LSRPLVPLPF SSNGKLSRSS FLTVGSSSGL YTSQKLGKPN ILRSLPKLGR RFQTVPEDEM 780 PEILCGDSSP STGIKSASPN SKRVSPPHGD FGSSPSRRSG RKLILQSIPS FPSLTPKH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 2e-17 | 547 | 667 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 2e-17 | 547 | 667 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
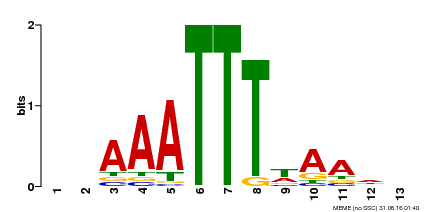
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018818657.1 | 0.0 | PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X3 | ||||
| TrEMBL | A0A2I4EGY1 | 0.0 | A0A2I4EGY1_JUGRE; protein tesmin/TSO1-like CXC 2 isoform X3 | ||||
| STRING | XP_008224266.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2619 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22780.1 | 1e-135 | Tesmin/TSO1-like CXC domain-containing protein | ||||



