 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | WALNUT_00013305-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 302aa MW: 33492 Da PI: 4.9258 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 29.9 | 1.2e-09 | 141 | 181 | 3 | 43 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNk 43
+k+ +r +NR AA +sR+RKk+++ Le k+ Le+e
WALNUT_00013305-RA 141 LSKKRKRQLRNRDAAVKSRERKKMYVRDLEMKSRYLEGECL 181
57999********************************9974 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 6.9E-11 | 137 | 199 | No hit | No description |
| SMART | SM00338 | 0.0016 | 139 | 203 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.278 | 141 | 183 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.7E-8 | 142 | 185 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.25E-10 | 143 | 199 | No hit | No description |
| CDD | cd14704 | 1.20E-17 | 144 | 195 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 146 | 161 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MEDPEFLVED DIIGRINFDG LFDEFPDGTN LFSVELPVFT DSSSAEAFLK PSPDSGSSWI 60 DEIETLLMKE DDGRIVAVEP TKEFCDSFLA DIIVDNSPGE GSSGADKDSS DCSDGGNGNC 120 DKGKFSNDNH GDDQNDADDP LSKKRKRQLR NRDAAVKSRE RKKMYVRDLE MKSRYLEGEC 180 LRLGRLLQCC YAENQALRLS LHSGSAFGAS VTKQESAVLL LESLLLGSLL WFLGITCLFT 240 LPTMAQSILK RAPLEDVGKK NPGVALRGAR SKMFGYLVVQ SFVKSRRCKA SRMKMKPNYL 300 II |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 143 | 162 | KRKRQLRNRDAAVKSRERKK |
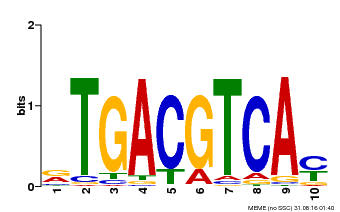
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018843068.1 | 0.0 | PREDICTED: bZIP transcription factor 60-like | ||||
| TrEMBL | A0A2I4GGP0 | 0.0 | A0A2I4GGP0_JUGRE; bZIP transcription factor 60-like | ||||
| STRING | XP_002510740.1 | 1e-85 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12009 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 6e-14 | basic region/leucine zipper motif 60 | ||||



